In a typical data science workflow there are multiple steps involved from data aggregation, cleaning, exploratory analysis, modeling and so on. As the data science community matures, we are seeing that there are a variety of languages which provide better capabilities for specific steps in the data science workflow. R is typically used for data transformations, statistical models, and visualizations, while Python provides more robust functions for machine learning. In addition to this, Spark provides an environment to process high volume data - both as one-time/ batch or as streams.
The job of today’s data scientist is changing from one where they are married to a specific tool or language, to one where they are using all these tools for their specialized purposes. The key problem then becomes one of translation between these tools for seamless analysis. Additionally, in the work of a data scientist, there is a need to perform the same task repeatedly, as well as put certain analysis flows (or) pipelines into production to work on new data periodically, or work on streaming data.
Recently, interfaces for using these various tools have been published. In terms of R packages, the reticulate package provides an interface to Python, and the SparkR and sparklyr packages provide an interface to Spark.
The analysisPipelines package uses these interfaces to enable Interoperable Pipelines i.e. the ability compose and execute a reusable data science pipeline which can contain functions to be executed in an R environment, in a Python environment or in a Spark environment. These pipelines can saved and loaded, to enable batch operation as datasets get updated with new data.
The goal of the analysisPipelines package is to make the job of the data scientist easier and help them compose pipelines of analysis which consist of data manipulation, exploratory analysis & reporting, as well as modeling steps. The idea is for data scientists to use tools of their choice through an R interface, using this package Essentially, it allows data scientists to:
This package supports for both batch/ repeated pipelines, as well as streaming pipelines.
For batch pipelines, the vision is to enable interoperable pipelines which execute efficiently with functions in R, Spark and Python
For streaming pipelines, the package allows for streaming analyses through Apache Spark Structured Streaming.
The analysisPipelines package uses S4 classes and methods to implement all the core functionality. The fundamental class exposed in this package is the BaseAnalysisPipeline class on which most of the core functions are implemented. The user, however, interacts with the AnalysisPipeline and StreamingAnalysisPipeline classes for batch and streaming analysis respectively.
The package stays true to the tidyverse pipelining style which also fits nicely into the idea of creating pipelines. The core mechanism in the package is to instantiate a pipeline with data and then pipeline required functions to the object itself.
The package allows both the use of magrittr pipe (%>%) or the pipeR pipe (%>>%).
As of this version, the package supports functions executed on R, or Spark through the SparkR interface, as well as Python functions run through reticulate for batch pipelines. It also supports Apache Spark Structured Streaming pipelines for streaming analyses.
This package contains 7 vignettes:
When the package is installed and loaded, vignettes 1 & 7 have all the chunks evaluated. Other vignettes require specific Python and Spark configurations and hence all chunks are not evaluated as part of the package. However, an evaluated version of vignettes 2-5 can be found in the knit-vignettes folder in the Github project. Vignette 6 is a shiny document which can be run.
An object of class AnalysisPipeline can be created like so:
{r creating object, warning=F} obj <- AnalysisPipeline(input = iris) class(obj)
While initializing the object, an input dataframe can be provided on which the pipeline should work, either by providing the filePath to a .csv file through the filePath argument, or by providing R dataframe available in the session, through the input argument
The AnalysisPipeline object has a set of getters, for retrieving various slots containing data and metadata required for pipeline execution. The most basic of them is the getInput method which retrieves the input dataframe with which the object has been initialized. If not initialized with a dataframe, the setInput method can be used to do so.
{r printing object contents, warning=F} obj %>>% getInput %>>% str getRegistry()
The getRegistry function retrieves the set of functions and their metadata available for pipelining. Any AnalysisPipeline object comes with a set of pre-registered functions which can be used out-of-the-box. Of course, the user can register her own functions, to be used in the pipeline. We will explore this later on.
There are two types of functions which can be pipelined:
Both pre-registered and user-defined functions work with the AnalysisPipeline object in the same way i.e. regardless of who writes the function, they follow the same semantics.
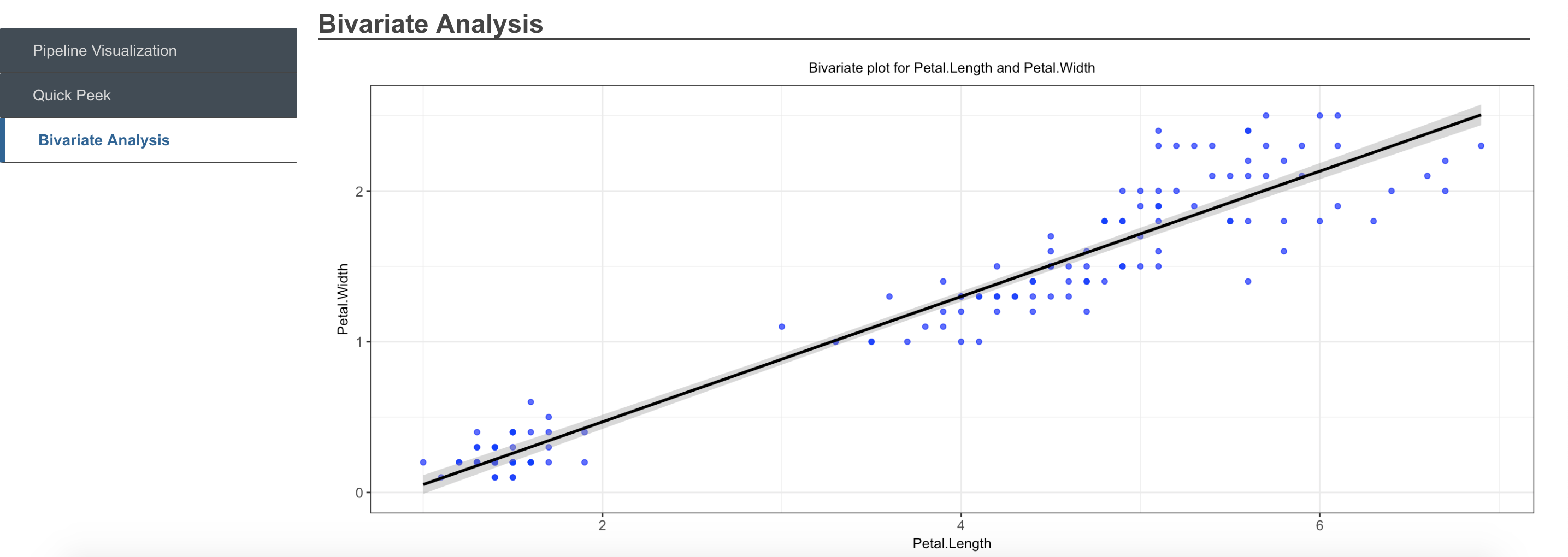
We’ll now take a look at creating a simple pipeline, with some of the pre-registered functions available in the registry. We pipeline the univarCatDistPlots function (available as a pre-registered utility function,which generates a chart showing distribution of a categorical variable in a dataset), by simply using the pipe or double pipe operator, and providing the required additional parameters apart from the data on which it needs to operate, as we have already initialized the AnalysisPipeline object with the data.
Note that unless assigned to the same or another object, the pipeline does not get stored.
We can access the details of the pipeline as a tibble through the getPipeline method.
{r pipe demo 1, warning=F} # Running univariate categorical distribution plot on the constructed object # ?analysisPipelines::univarCatDistPlots obj1 <- obj %>>% univarCatDistPlots(uniCol = "Species", priColor = "blue", optionalPlots = 0, storeOutput = T) obj1 %>>% getPipeline
You can register your own data or non-data functions by calling registerFunction. This adds the user-defined function to the registry. The registry is maintained by the package and once registered, functions can be used across pipeline objects. The registry can be view by calling the getRegistry function.
In order to register a function, first the function must be defined in the Global environment, before calling registerFunction.
bivariatePlots <- function(dataset, select_var_name_1, select_var_name_2,
priColor = "blue", secColor='black') {
x=dataset[, select_var_name_1]
y=dataset[, select_var_name_2]
bivarPlot <- ggplot2::ggplot(dataset, ggplot2::aes(x,y)) +
ggplot2::geom_point(color=priColor,alpha=0.7) +
ggplot2::geom_smooth(method = lm,color=secColor) +
ggplot2::xlab(select_var_name_1) +
ggplot2::ylab(select_var_name_2) +
ggplot2::theme_bw() +
ggplot2::ggtitle(paste('Bivariate plot for', select_var_name_1,
'and', select_var_name_2, sep=' ')) +
ggplot2::theme(plot.title = ggplot2::element_text(hjust = 0.5, size = 10),
axis.text = ggplot2::element_text(size=10),
axis.title=ggplot2::element_text(size=10))
return(bivarPlot)
}
registerFunction(functionName = "bivariatePlots", heading = "Bivariate Analysis")Now the newly registered user-defined function can be used as part of the pipeline, exactly as described before. For example, we add it to a pipeline which already contains some functions. The function then gets added to the end of the pipeline
```{r register function 2, warning=F} # Chaining the user-defined function to the object’s pipeline where it was registered obj2 <- obj2 %>>% bivariatePlots(select_var_name_1 = ‘Sepal.Length’, select_var_name_2 = ‘Sepal.Width’, priColor = “blue”, secColor = “black”)
obj2 %>>% getPipeline
## Complex pipelines and formula semantics
In addition to simple linear pipelines, more complex pipelines can also be defined. There are cases when the outputs of previous functions in the pipeline, as inputs to arbitrary parameters of subsequent functions.
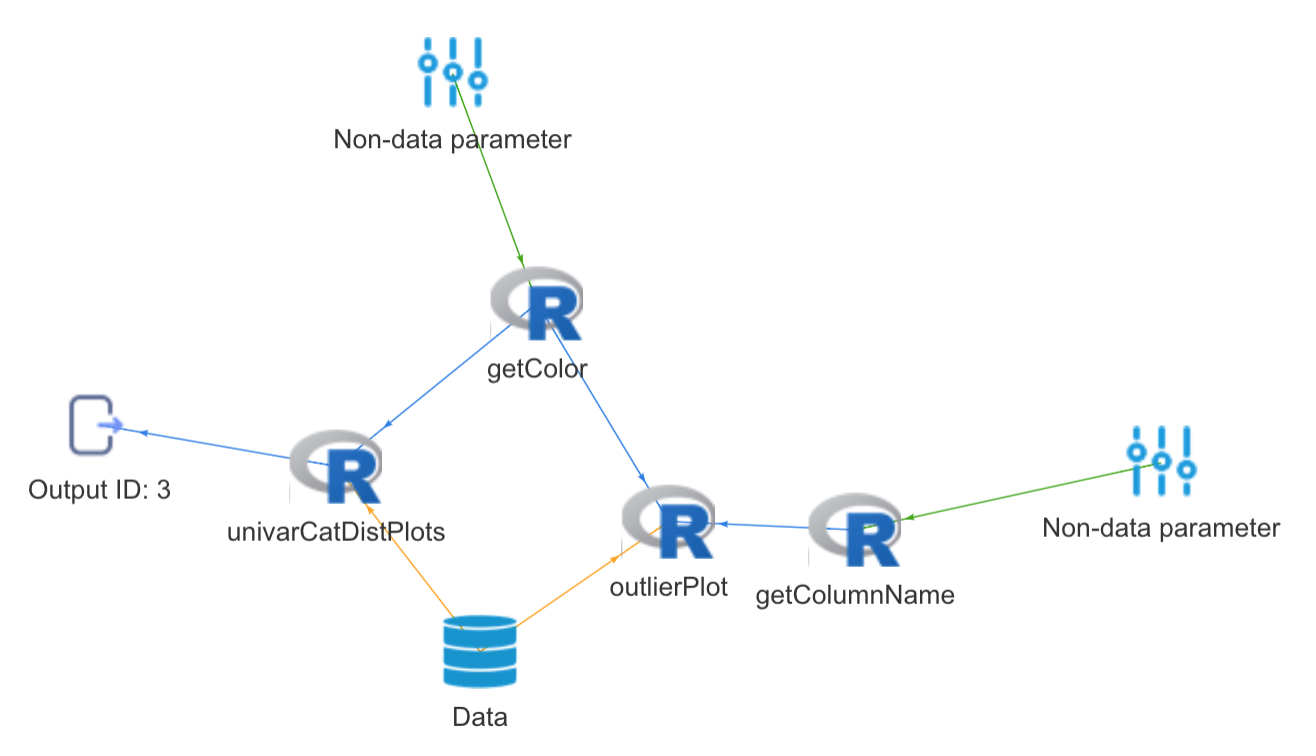
The package defines certain *formula* semantics to accomplish this. We take the example of two simple user-defined functions, both which simply return the color of the graph, as well as the column on which the graph should be plotted, in order to illustrate how this works.
Preceding outputs can be passed to subsequent functions simply by specifying a **formula** of the form 'f*id*' against the argument to which the output is to be passed . The ID represents the ID of the function in the pipeline. For example, to pass the output of function with ID '1' as an argument to a parameter of a subsequent function, the formula '~f1' is passed to that corresponding argument.
```r
obj %>>% getColor(color = "blue") %>>% getColumnName(columnName = "Sepal.Length") %>>%
univarCatDistPlots(uniCol = "Species", priColor = ~f1, optionalPlots = 0, storeOutput = T) %>>%
outlierPlot(method = "iqr", columnName = ~f2, cutoffValue = 0.01, priColor = ~f1 , optionalPlots = 0) -> complexPipeline
complexPipeline %>>% getPipeline
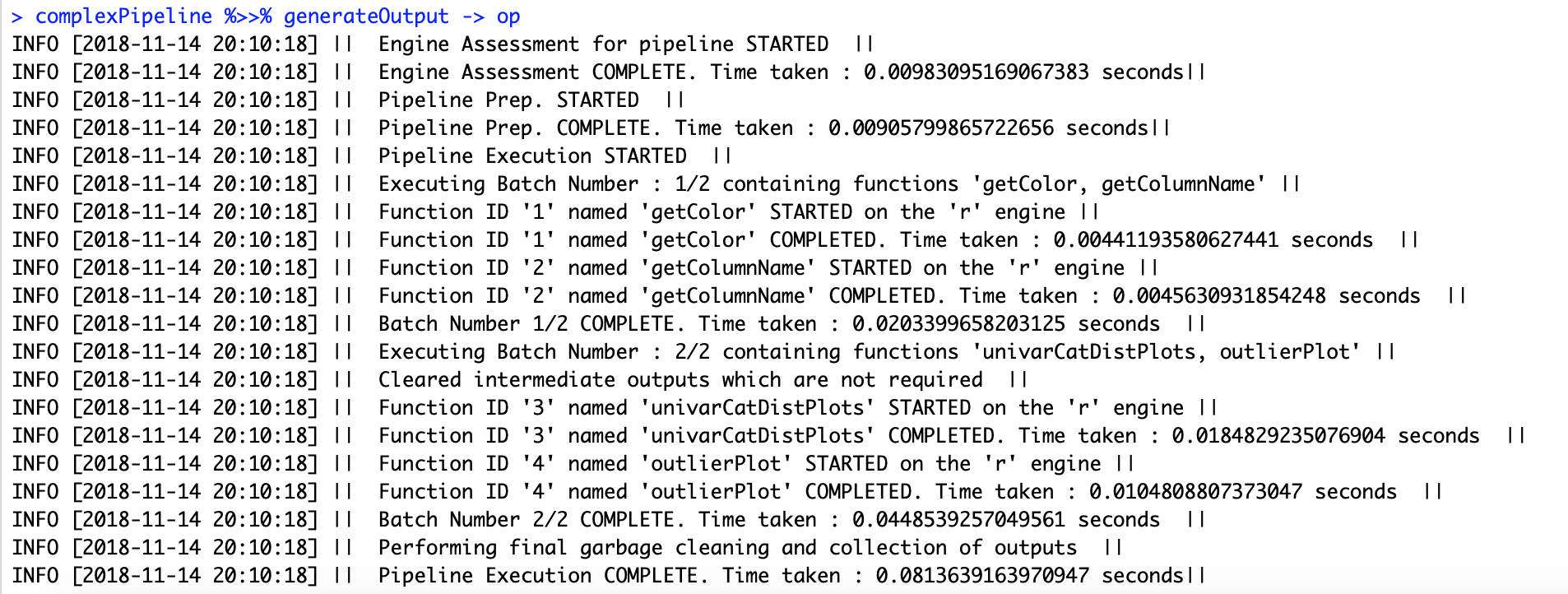
complexPipeline %>>% generateOutput -> op
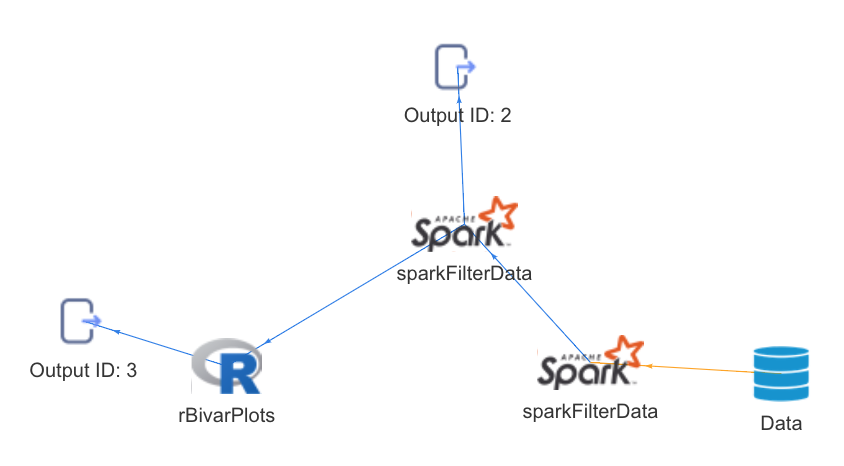
op %>>% getOutputById("4")Interoperable pipelines containing functions operating on different engines such as R, Spark and Python can be configured and executed through the analysisPipelines package. Currently, the package supports interoperable pipelines containing R and Spark batch functions.

Pipelines can be visualized as directed graphs, providing information about the engines being used, function dependencies and so on.
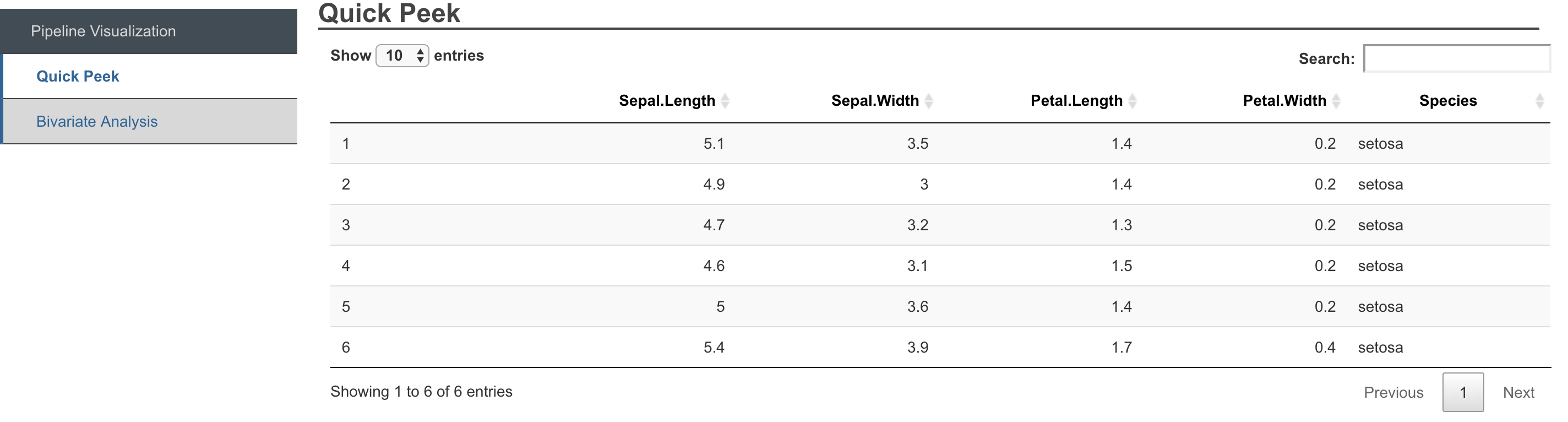
Outputs generated from pipelines can easily be exported to formatted reports, showcasing the results, generating pipeline as well as a peek at the data
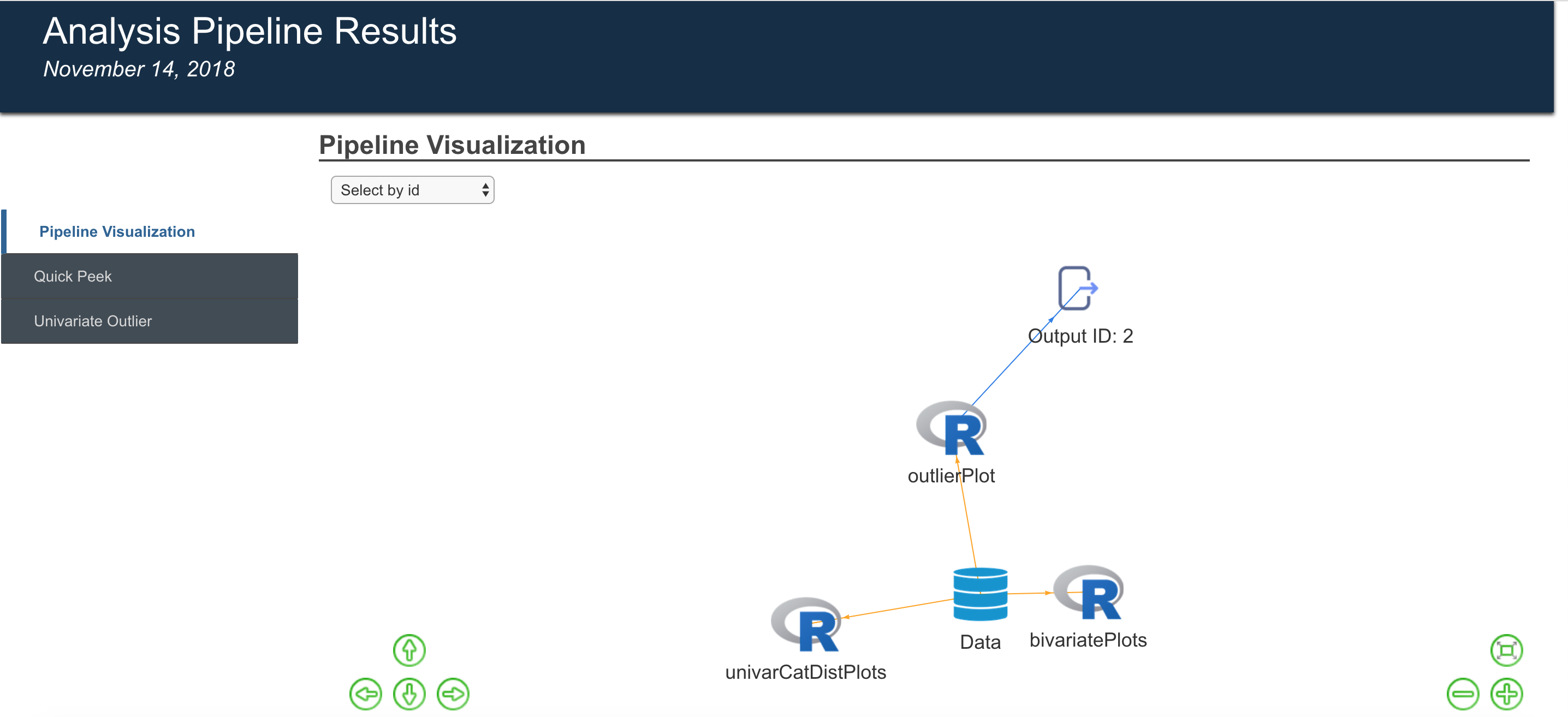
The meta-pipeline construct is one which allows users to export pipelines they have created for a particular use case to a general analysis flow which can be used for a different dataset and different set of parameters. A pipeline is one where the data can change, though retaining the same schema, and the same set of parameters for the functions. A meta-pipeline is one where only the analysis flow, function dependencies and so on are retained. The specific parameters for each of the functions can be set differently for a new use case.
The objective of a meta-pipeline is to define and execute reusable analysis flows. They can be used to: * Document best practices for a particular problem * Templatize analyses for particular situations
Meta-pipelines can be created by exporting from pipelines, and new pipelines instantiated from a meta-pipeline, with an easy-to-use method to set the new values of parameters.
The ‘analysisPipelines’ package internally converts the pipeline defined by the user into a directed graph which captures dependencies of each function in the pipeline on data, other arguments as well as outputs as other functions.
When it is required to generate the output, the pipeline is first prepped by performing a topological sort of the directed graph, and identifying sets (or) batches of independent functions and a sequence of batches for execution. A later release of the package will allow for parallel execution of these independent functions
Memory is managed efficiently, by only storing outputs which the user has explicitly specified, or temporarily storing intermediate outputs required for subsequent functions only until they are required for processing. Garbage cleaning is performed after the execution of each batch in order to manage memory effectively.
In the case of Interoperable pipelines executing across multiple engines such as R, Spark and Python, type conversions between data types in the different engines is minimized by identifying the optimal number of type conversions, before execution starts
The package provides logging capabilities for execution of pipelines, as you might have noted when the output was generated in sections above. By default, logs are written to the console, but alternatively the user can specify an output file to which the logs need to be written through the setLoggerDetails function.
Logs capture errors, as well as provide information on the steps being performed, execution times and so on.
By default, when a function is registered, a generic exception handling function which captures the R error message, in case of error is registered against each function in the registry. The user can define a custom exception handling function, by defining it and providing it during the time of registration. The function should take 1 argument, which is the error object.