
You can get the development version from GitHub:
if (!require("remotes")) install.packages("remotes")
remotes::install_github("toledo60/ggDoE")With ggDoE you’ll be able to generate common plots used in Design of Experiments with ggplot2.
library(ggDoE)The following plots are currently available:
Alias Matrix
Correlation matrix plot to visualize the Alias matrix
alias_matrix(design=aliased_design)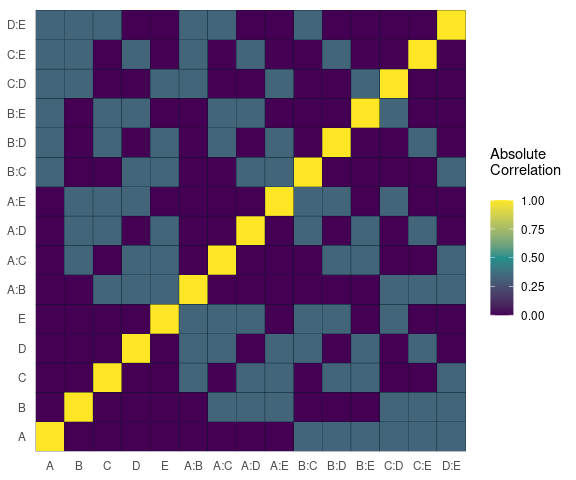
Box-Cox Transformation
model <- lm(s2 ~ (A+B+C+D),data = adapted_epitaxial)
boxcox_transform(model,lambda = seq(-5,5,0.2))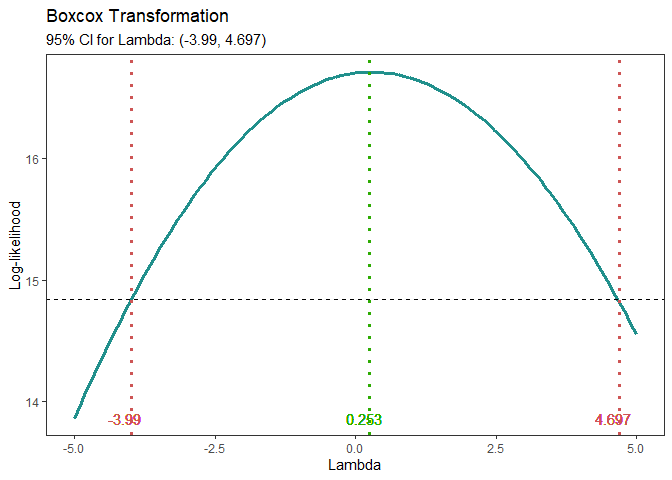
Lambda Plot
Obtain the trace plot of the t-statistics after applying Boxcox transformation across a specified sequence of lambda values
model <- lm(s2 ~ (A+B+C)^2,data=original_epitaxial)
lambda_plot(model)
lambda_plot(model, lambda = seq(0,2,by=0.1))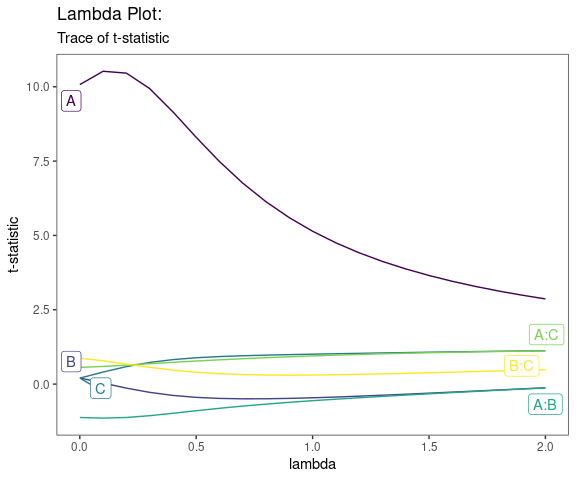
Boxplots
data <- ToothGrowth
data$dose <- factor(data$dose,levels = c(0.5, 1, 2),
labels = c("D0.5", "D1", "D2"))
gg_boxplots(data,response = 'len',
factor = 'dose')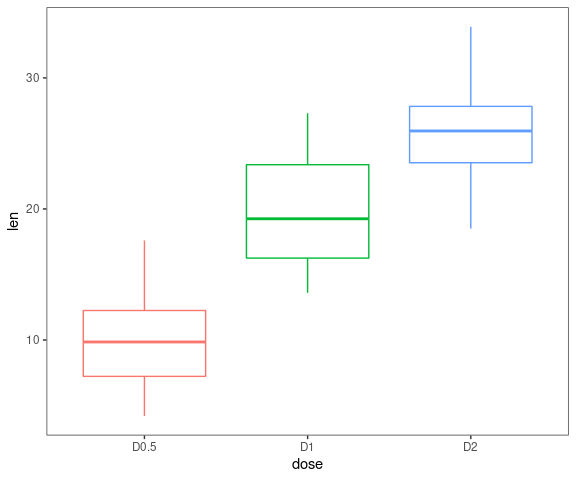
gg_boxplots(data,response = 'len',
factor = 'dose',
group_var = 'supp',
color_palette = 'viridis',
jitter_points = TRUE)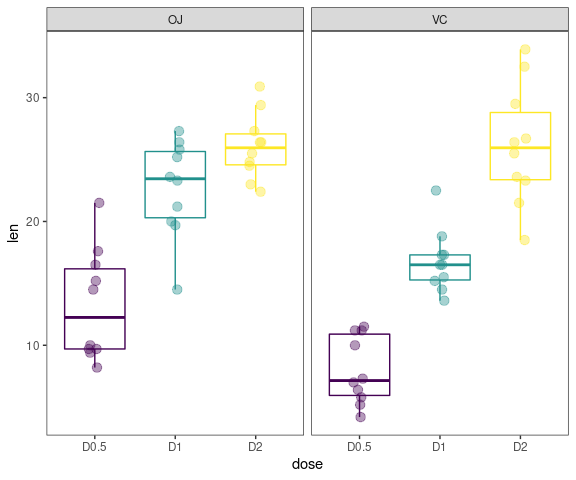
Regression Diagnostic Plots
The default plots are 1-4
model <- lm(mpg ~ wt + am + gear + vs * cyl, data = mtcars)
diagnostic_plots(model,which_plots=1:6)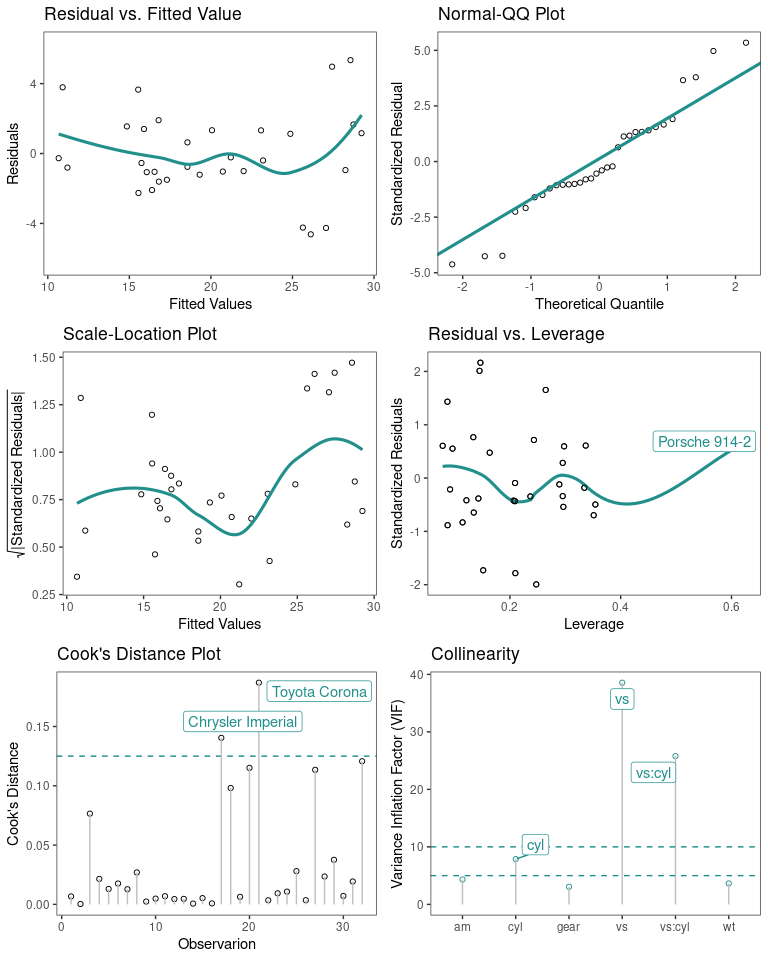
Half-Normal Plot
model <- lm(ybar ~ (A+B+C+D)^4,data=adapted_epitaxial)
half_normal(model)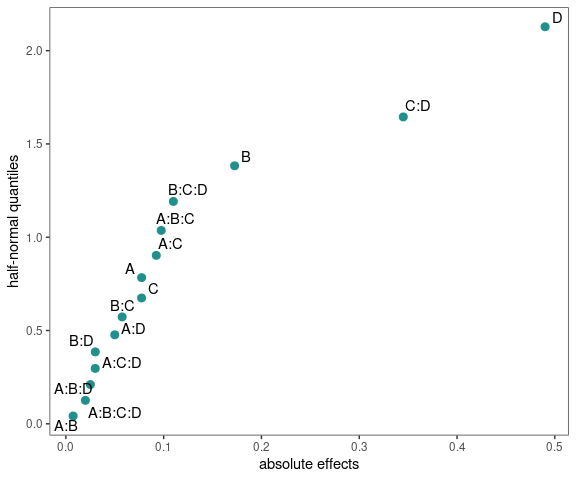
half_normal(model,method='Zahn',alpha=0.1,
ref_line=TRUE,label_active=TRUE,
margin_errors=TRUE)
Interaction Effects Plot (Factorial Design)
Interaction effects plot between two factors in a factorial design
interaction_effects(adapted_epitaxial,response = 'ybar',
exclude_vars = c('s2','lns2'))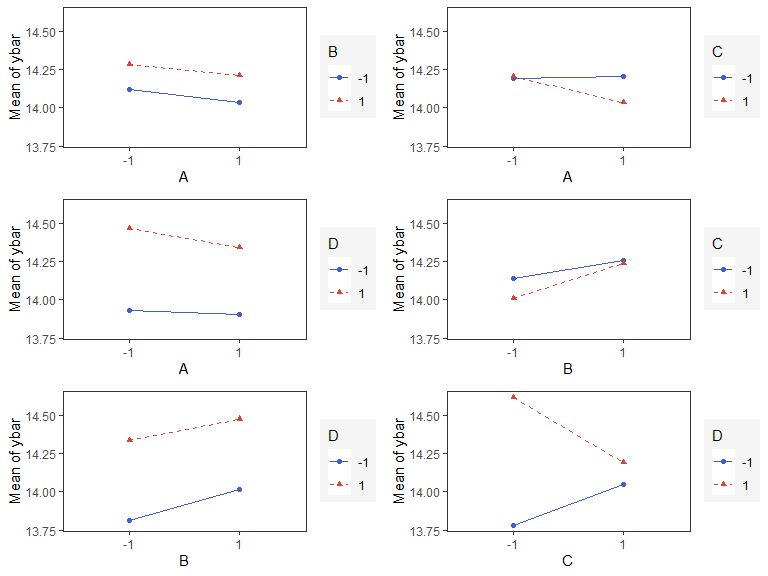
interaction_effects(adapted_epitaxial,response = 'ybar',
exclude_vars = c('A','s2','lns2'),
n_columns=3)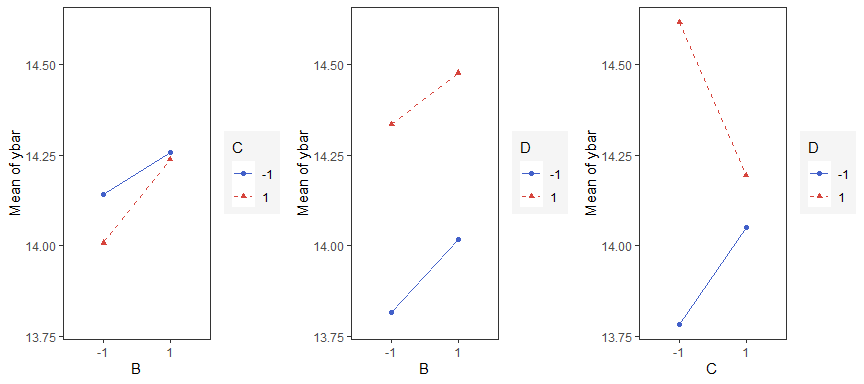
Main Effects Plots (Factorial Design)
Main effect plots for each factor in a factorial design
main_effects(original_epitaxial,
response='s2',
exclude_vars = c('ybar','lns2'))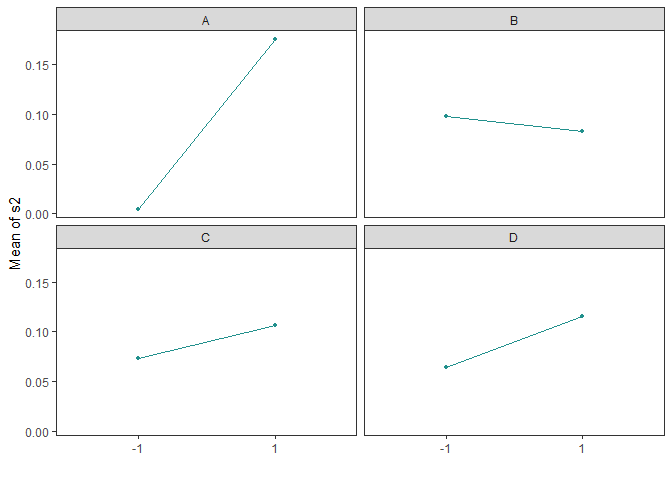
main_effects(original_epitaxial,
response='s2',
exclude_vars = c('A','ybar','lns2'),
color_palette = 'viridis',
n_columns=3)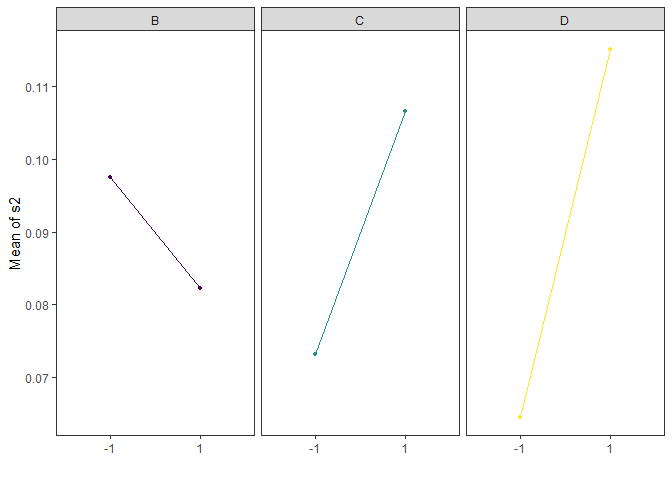
Contour Plots
contour plot(s) that display the fitted surface for an rsm object involving two or more numerical predictors
heli.rsm <- rsm::rsm(ave ~ SO(x1, x2, x3, x4),
data = rsm::heli)gg_rsm(heli.rsm,form = ~x1+x2+x3+x4,
at = rsm::xs(heli.rsm))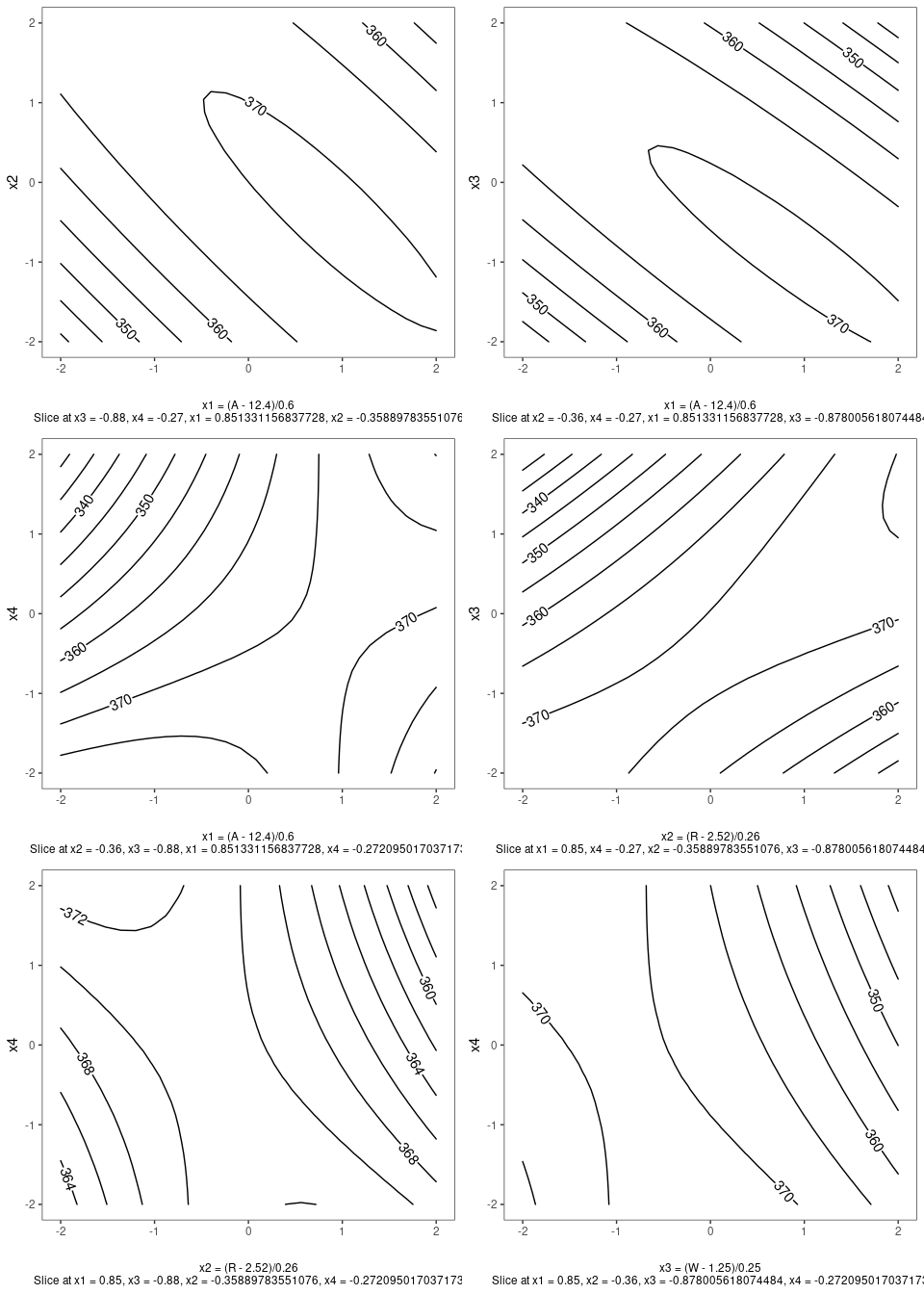
gg_rsm(heli.rsm,form = ~x1+x2+x3+x4,
at = rsm::xs(heli.rsm),
filled = TRUE)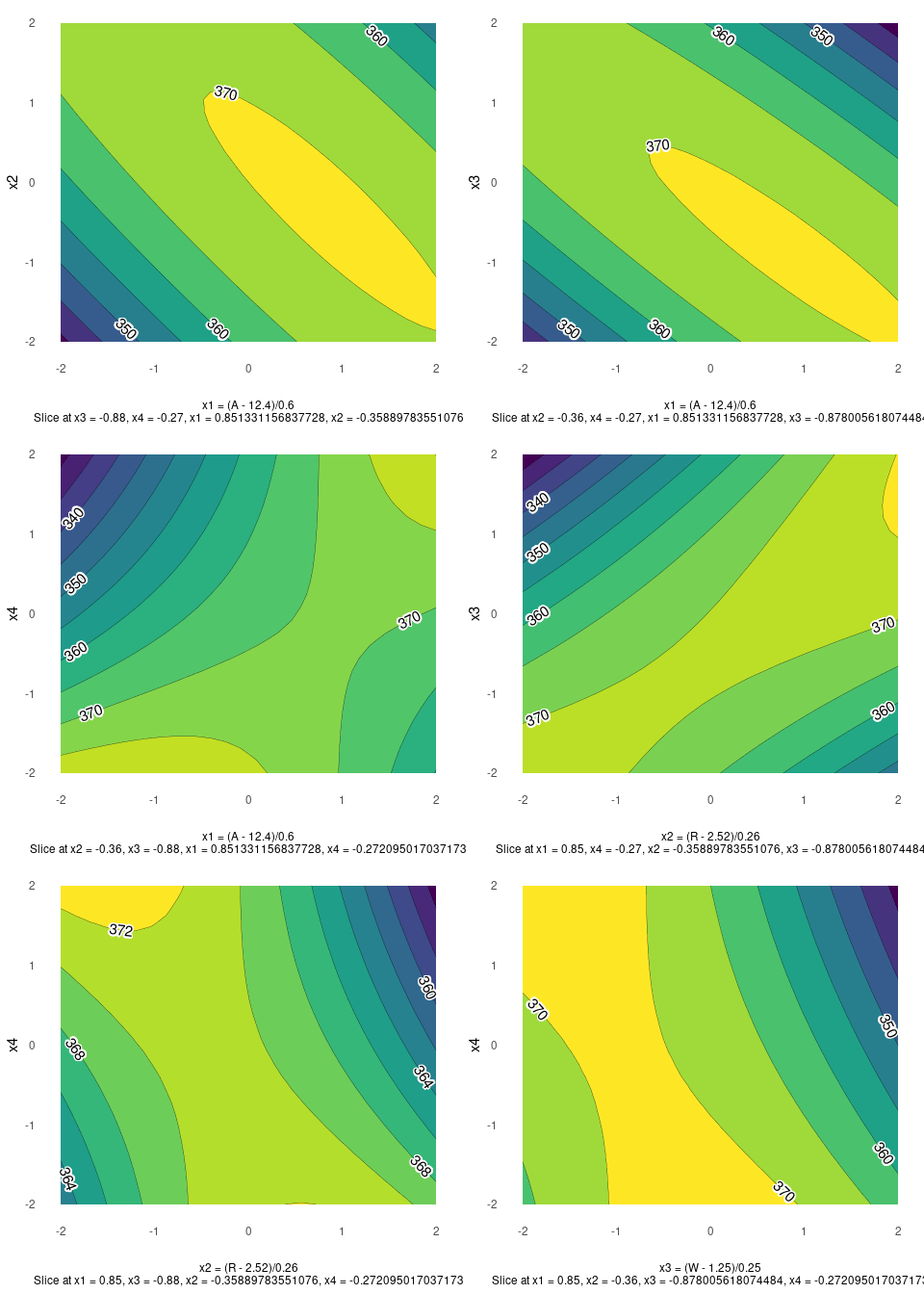
Pareto Plot
Pareto plot of effects with cutoff values for the margin of error (ME) and simultaneous margin of error (SME)
model <- lm(lns2 ~ (A+B+C+D)^4,data=original_epitaxial)
pareto_plot(model)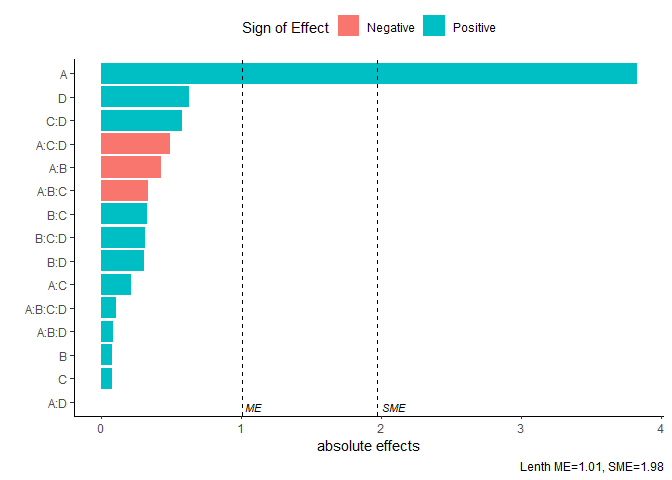
pareto_plot(model,method='Zahn',alpha=0.1)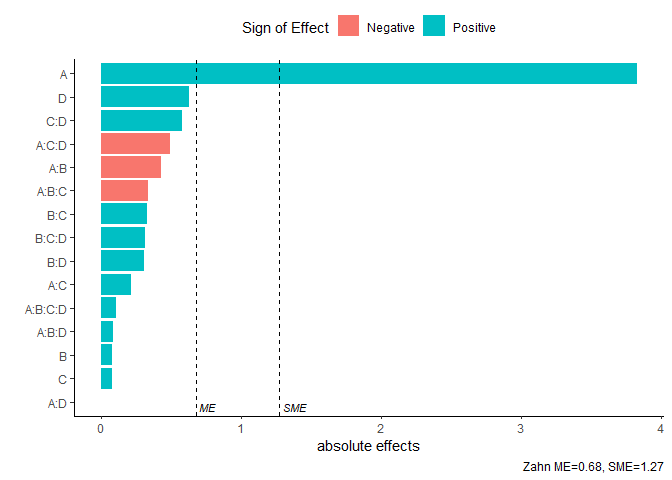
Two Dimensional Projections
This function will output all two dimensional projections from a Latin hypercube design
set.seed(10)
X <- lhs::randomLHS(n=10, k=4)
twoD_projections(X,n_columns=3,grid = TRUE)
Lastly, the following datasets/designs are included in ggDoE as tibbles:
adapted_epitaxial: Adapted epitaxial layer
experiment obtain from the book
“Experiments: Planning,
Analysis, and Optimization, 2nd Edition”
original_epitaxial: Original epitaxial layer
experiment obtain from the book
“Experiments: Planning,
Analysis, and Optimization, 2nd Edition”
aliased_design: D-efficient minimal aliasing
design obtained from the article
“Efficient Designs With
Minimal Aliasing by Bradley Jones and Christopher J. Nachtsheim”
Source: https://www.tandfonline.com/doi/abs/10.1198/TECH.2010.09113
If you want to cite this package in a scientific journal or in any
other context, run the following code in your R console
citation('ggDoE')Warning in citation("ggDoE"): no date field in DESCRIPTION file of package
'ggDoE'
Warning in citation("ggDoE"): could not determine year for 'ggDoE' from package
DESCRIPTION file
To cite package 'ggDoE' in publications use:
Toledo Luna J (????). _ggDoE: Modern Graphs for Design of Experiments
with 'ggplot2'_. R package version 0.7.8,
<https://ggdoe.netlify.app>.
A BibTeX entry for LaTeX users is
@Manual{,
title = {ggDoE: Modern Graphs for Design of Experiments with 'ggplot2'},
author = {Jose {Toledo Luna}},
note = {R package version 0.7.8},
url = {https://ggdoe.netlify.app},
}Note: Once this package is submitted to CRAN the date warning will disappear. Simply change (????) to (2022)
I welcome feedback, suggestions, issues, and contributions! Check out the CONTRIBUTING file for more details.