
‘ggVennDiagram’ enables fancy Venn plot with 2-7 sets
and generates publication quality figure.
You can install the released version of ggVennDiagram from CRAN with (under evaluation in CRAN):
install.packages("ggVennDiagram")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("gaospecial/ggVennDiagram")If you find ggVennDiagram is useful and used it in academic papers, you may cite this package as:
Gao, C.-H., Yu, G., and Cai, P. (2021). ggVennDiagram: An Intuitive, Easy-to-Use, and Highly Customizable R Package to Generate Venn Diagram. Frontiers in Genetics 12, 1598. doi: 10.3389/fgene.2021.706907.
ggVennDiagram maps the fill color of each region to
quantity, allowing us to visually observe the differences between
different parts.
library(ggVennDiagram)
genes <- paste("gene",1:1000,sep="")
set.seed(20210419)
x <- list(A=sample(genes,300),
B=sample(genes,525),
C=sample(genes,440),
D=sample(genes,350))ggVennDiagram return a ggplot object, the
fill/edge colors can be further modified with ggplot
functions.
library(ggplot2)
ggVennDiagram(x) + scale_fill_gradient(low="blue",high = "red")
ggVennDiagram(x) + scale_color_brewer(palette = "Paired")
ggVennDiagram now support 2-7 dimension Venn plot. The
generated figure is generally ready for publish. The main function
ggVennDiagram() will check how many items in the first
parameter and call corresponding function automatically.
The parameter category.names is set names. And the
parameter label can label how many items are included in
each parts.
ggVennDiagram(x,category.names = c("Stage 1","Stage 2","Stage 3", "Stage4"))
ggVennDiagram(x,category.names = c("Stage 1","Stage 2","Stage 3", "Stage4"), label = "none")
Set label_alpha = 0 to remove label background
ggVennDiagram(x, label_alpha=0)
Note: you need to install the GitHub version to enable these functions.
We implemented the process_region_data() to get
intersection values.
y <- list(
A = sample(letters, 8),
B = sample(letters, 8),
C = sample(letters, 8),
D = sample(letters, 8)
)
process_region_data(Venn(y))
#> # A tibble: 15 × 5
#> component id item count name
#> <chr> <chr> <list> <int> <chr>
#> 1 region 1 <chr [1]> 1 A
#> 2 region 2 <chr [1]> 1 B
#> 3 region 3 <chr [3]> 3 C
#> 4 region 4 <chr [5]> 5 D
#> 5 region 12 <chr [3]> 3 A..B
#> 6 region 13 <chr [2]> 2 A..C
#> 7 region 14 <chr [0]> 0 A..D
#> 8 region 23 <chr [0]> 0 B..C
#> 9 region 24 <chr [0]> 0 B..D
#> 10 region 34 <chr [0]> 0 C..D
#> 11 region 123 <chr [1]> 1 A..B..C
#> 12 region 124 <chr [1]> 1 A..B..D
#> 13 region 134 <chr [0]> 0 A..C..D
#> 14 region 234 <chr [2]> 2 B..C..D
#> 15 region 1234 <chr [0]> 0 A..B..C..DIf only several items were included, intersections may also be viewed
interactively by plotly method (if you have two many items,
this is useless).
ggVennDiagram(y, show_intersect = TRUE)In web browser or RStudio, you will get:
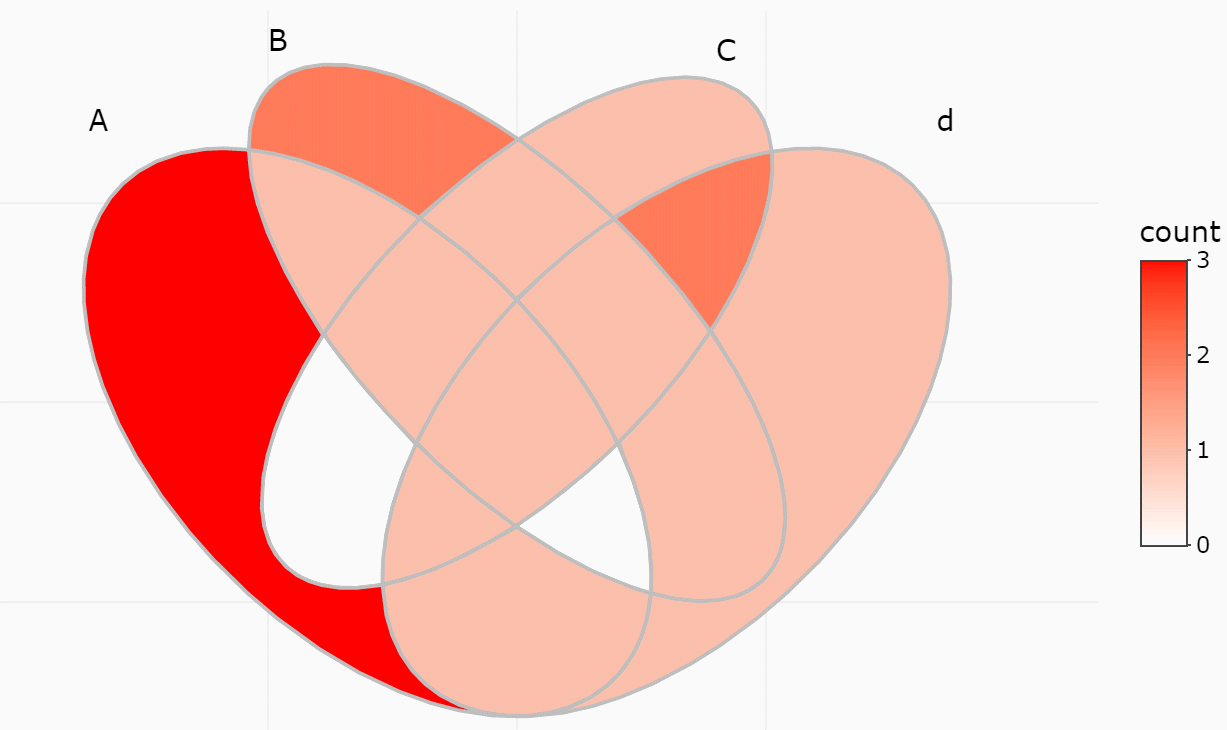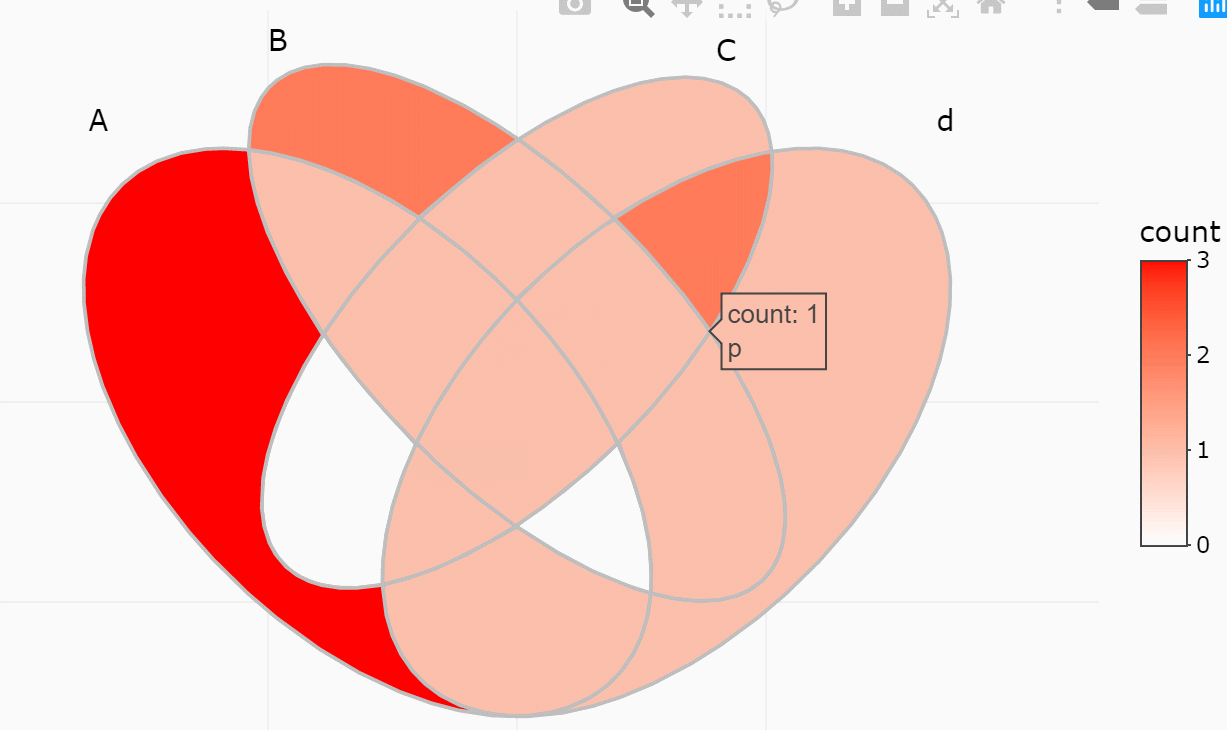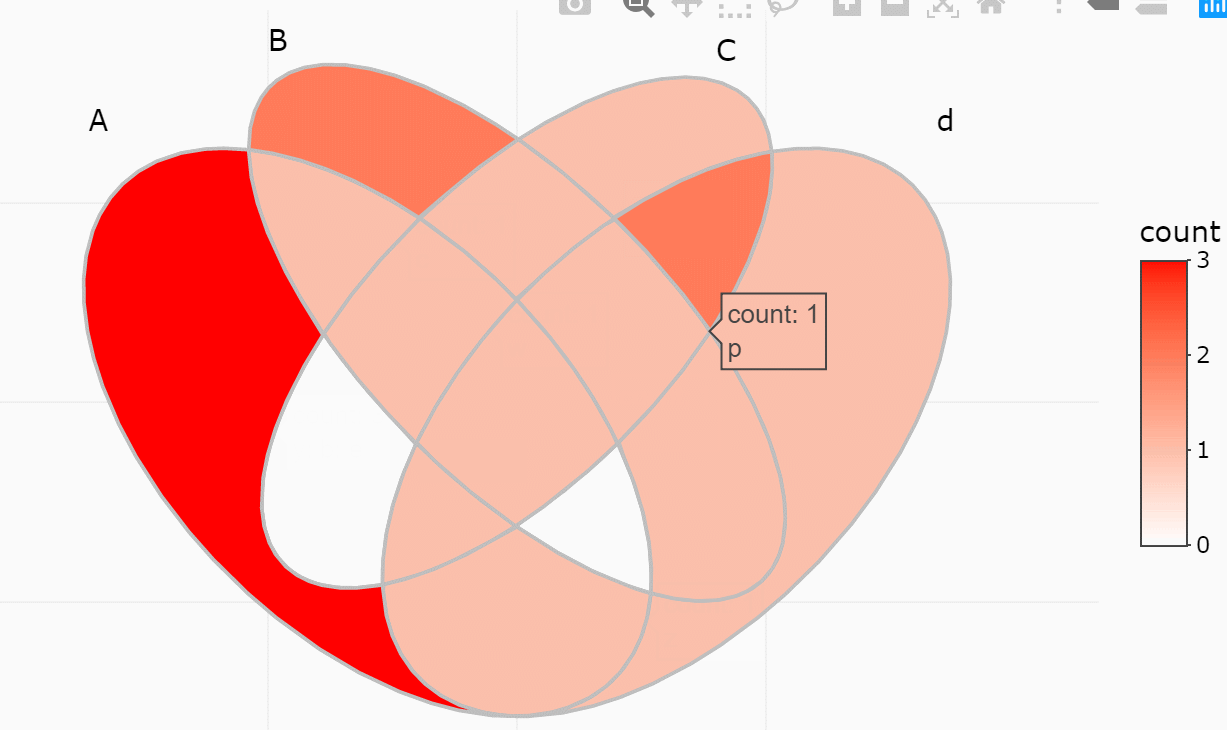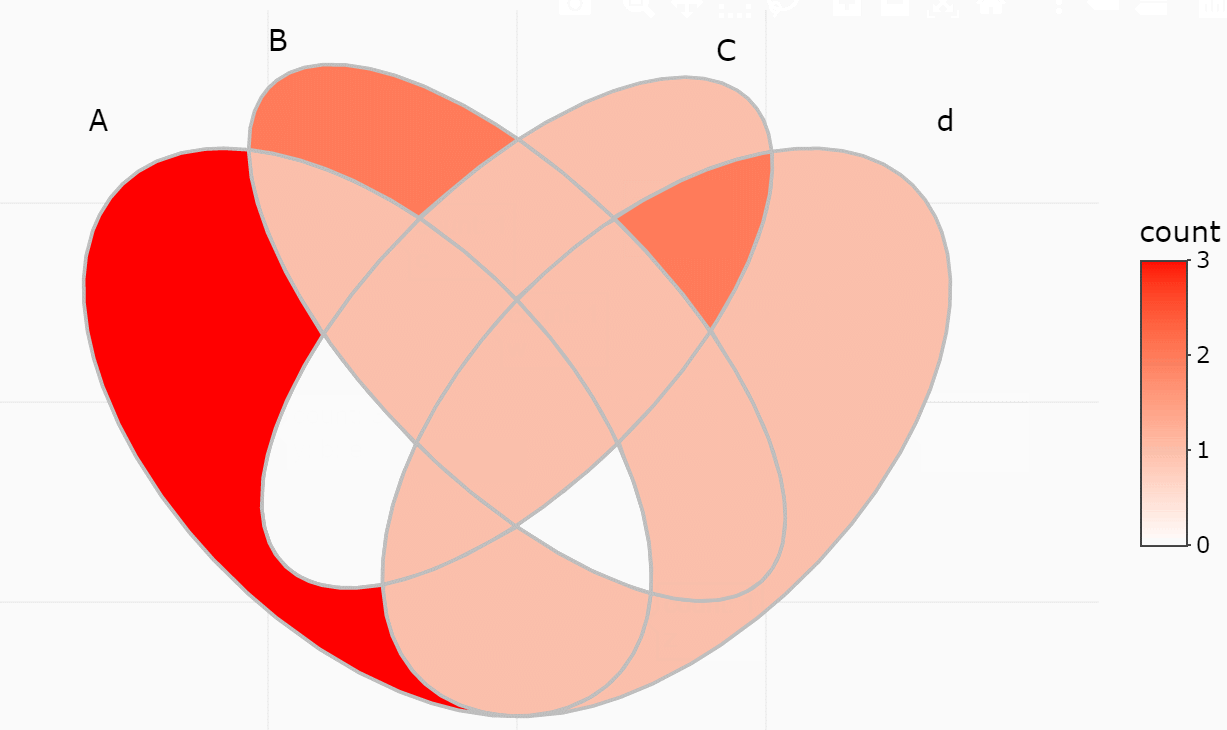
There are three components in a Venn plot: 1) the set labels; 2) the
edge of sets; and 3) the filling regions of each parts. We separately
stored these data in a structured S4 class VennPlotData
object, in which labels, edges and regions are stored as simple
features.
Simple features or simple feature access refers to a formal standard (ISO 19125-1:2004) that describes how objects in the real world can be represented in computers, with emphasis on the spatial geometry of these objects. But here we employed this to describe the coordinates of Venn components.
In general, ggVennDiagram() plot a Venn in three
steps:
shapes datasets.PlotDataVenn object that
includes all necessary definitions. We implement a number of set
operations functions to do this job.ggplot2 functions.venn <- Venn(x)
data <- process_data(venn)
data
#> An object of class "VennPlotData"
#> Slot "setEdge":
#> Simple feature collection with 4 features and 5 fields
#> Geometry type: LINESTRING
#> Dimension: XY
#> Bounding box: xmin: 0.0649949 ymin: 0.1849949 xmax: 0.9350051 ymax: 0.8391534
#> CRS: NA
#> # A tibble: 4 × 6
#> id geometry compo…¹ item count name
#> <chr> <LINESTRING> <chr> <nam> <int> <chr>
#> 1 1 (0.1025126 0.7174874, 0.09412107 0.7081191, 0… setEdge <chr> 300 A
#> 2 2 (0.2525126 0.8174874, 0.246341 0.8103391, 0.2… setEdge <chr> 525 B
#> 3 3 (0.7333452 0.8033452, 0.7262248 0.8095447, 0.… setEdge <chr> 440 C
#> 4 4 (0.8974874 0.7174874, 0.8881191 0.7258789, 0.… setEdge <chr> 350 D
#> # … with abbreviated variable name ¹component
#>
#> Slot "setLabel":
#> Simple feature collection with 4 features and 3 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 0.08 ymin: 0.78 xmax: 0.93 ymax: 0.86
#> CRS: NA
#> # A tibble: 4 × 4
#> id geometry component name
#> <chr> <POINT> <chr> <chr>
#> 1 1 (0.08 0.78) setLabel A
#> 2 2 (0.26 0.86) setLabel B
#> 3 3 (0.71 0.85) setLabel C
#> 4 4 (0.93 0.78) setLabel D
#>
#> Slot "region":
#> Simple feature collection with 15 features and 5 fields
#> Geometry type: POLYGON
#> Dimension: XY
#> Bounding box: xmin: 0.0649949 ymin: 0.1849949 xmax: 0.9350051 ymax: 0.8391534
#> CRS: NA
#> # A tibble: 15 × 6
#> id geometry compo…¹ item count name
#> <chr> <POLYGON> <chr> <lis> <int> <chr>
#> 1 1 ((0.1025126 0.7174874, 0.1118809 0.7258789, … region <chr> 41 A
#> 2 2 ((0.2525126 0.8174874, 0.2596609 0.823659, 0… region <chr> 151 B
#> 3 3 ((0.7333452 0.8033452, 0.7395447 0.7962248, … region <chr> 91 C
#> 4 4 ((0.8974874 0.7174874, 0.9058789 0.7081191, … region <chr> 75 D
#> 5 12 ((0.2494531 0.7508377, 0.266399 0.7472201, 0… region <chr> 49 A..B
#> 6 13 ((0.3598131 0.3161471, 0.3466157 0.3144203, … region <chr> 51 A..C
#> 7 14 ((0.6341476 0.306281, 0.6321686 0.2919527, 0… region <chr> 36 A..D
#> 8 23 ((0.4087951 0.6905086, 0.4240163 0.7044756, … region <chr> 111 B..C
#> 9 24 ((0.7013464 0.5605964, 0.7121743 0.5437184, … region <chr> 63 B..D
#> 10 34 ((0.7562978 0.7359764, 0.7555797 0.7233843, … region <chr> 53 C..D
#> 11 123 ((0.4254307 0.668526, 0.4425419 0.6552909, 0… region <chr> 49 A..B…
#> 12 124 ((0.6020164 0.4567956, 0.6098817 0.4389429, … region <chr> 38 A..B…
#> 13 134 ((0.4966178 0.374675, 0.4805314 0.3646387, 0… region <chr> 21 A..C…
#> 14 234 ((0.5085786 0.6114214, 0.5243976 0.6266822, … region <chr> 49 B..C…
#> 15 1234 ((0.5066822 0.5956024, 0.5213246 0.5792878, … region <chr> 15 A..B…
#> # … with abbreviated variable name ¹componentNow we can custom this figure.
ggplot() +
geom_sf(aes(fill=count), data = venn_region(data)) +
geom_sf(size = 2, lty = "dashed", color = "grey", data = venn_setedge(data), show.legend = F) +
geom_sf_text(aes(label = name), data = venn_setlabel(data)) +
geom_sf_label(aes(label=id), fontface = "bold", data = venn_region(data)) +
theme_void()
If you have reviewed my codes, you may find it is easy to support Venn Diagram for more than four sets, as soon as you find a ideal parameter to generate more circles or ellipses in the plot. The key point is to let the generated ellipses have exactly one intersection for each combination.
From v1.0, ggVennDiagram can plot up to seven dimension
Venn plot. We would like to acknowledgment the author of package
venn, for his kind help on sharing the required shape
coordinates for this feature.
However, Venn Diagram for more than four sets may be meaningless in some conditions, as some parts may be omitted in such ellipses. Therefore, it is only useful in specific conditions. For example, if the set intersection of all group are extremely large, you may use several ellipses to draw a “flower” to show that.
x <- list(A=sample(genes,300),
B=sample(genes,525),
C=sample(genes,440),
D=sample(genes,350),
E=sample(genes,200),
F=sample(genes,150),
G=sample(genes,100))
# seven dimension Venn plot
ggVennDiagram(x)
# six dimension Venn plot
ggVennDiagram(x[1:6])
# five dimension Venn plot
ggVennDiagram(x[1:5])
# four dimension Venn plot
ggVennDiagram(x[1:4])
# three dimension Venn plot
ggVennDiagram(x[1:3])
# two dimension Venn plot
ggVennDiagram(x[1:2])
I have released a online book to introduce the development of
ggVennDiagram, it contains a chapter that compare many
different R packages in plotting a Venn. Refer to Venn Diagram cookbook in R.
在 @GuangchuangYu
的公众号下面,我投稿了两篇文章,介绍了 “ggVennDiagram”
包开发的始末。