


MAPpoly (v. 0.3.0) is an R package to construct genetic maps in autopolyploids with even ploidy levels. In its current version, MAPpoly can handle ploidy levels up to 8 when using hidden Markov models (HMM), and up to 12 when using the two-point simplification. When dealing with large numbers of markers (> 10,000), we strongly recommend using high-performance computation.
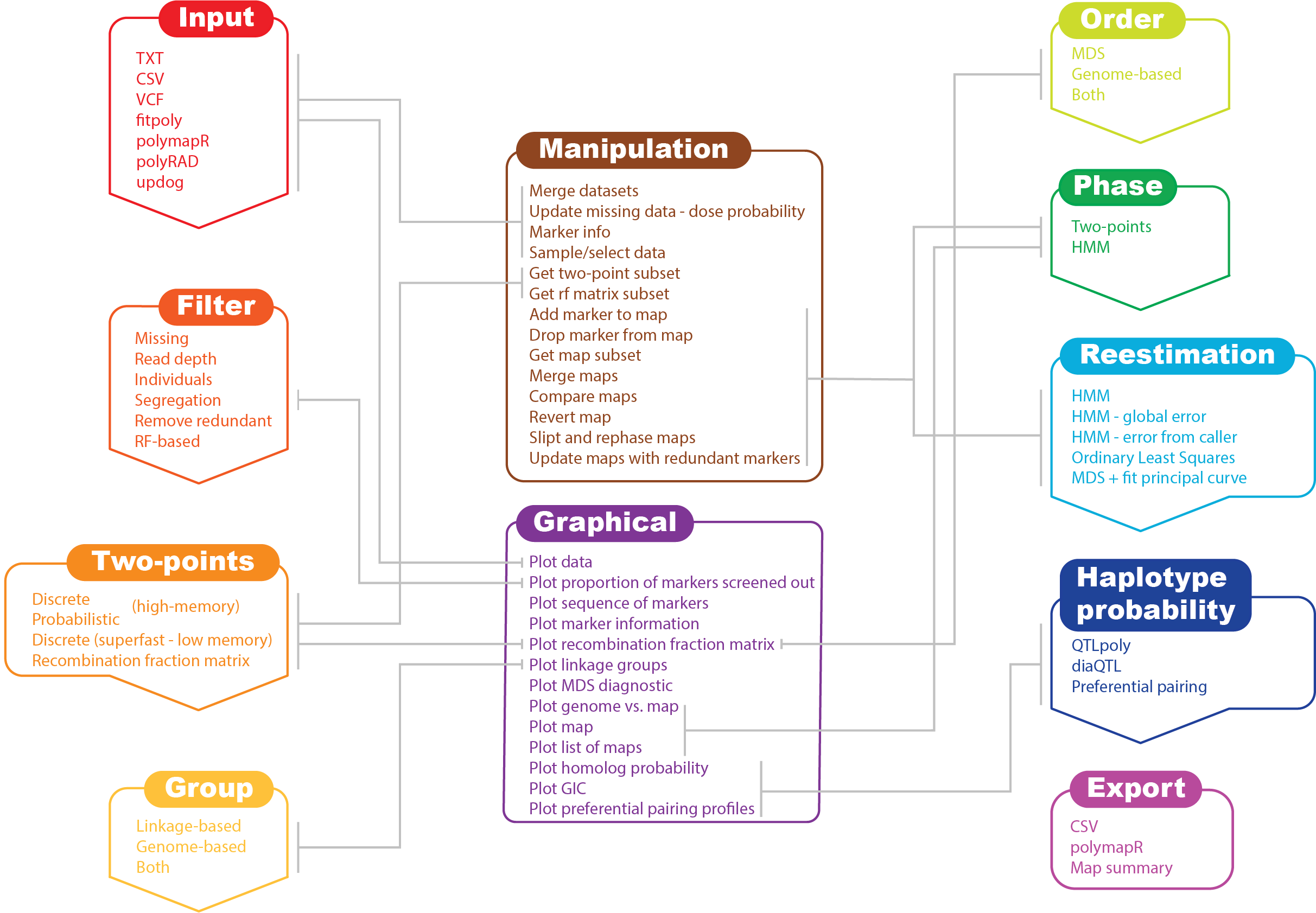
In its current version, MAPpoly can handle the following types of datasets:
MAPpoly also is capable of importing objects generated by the following R packages
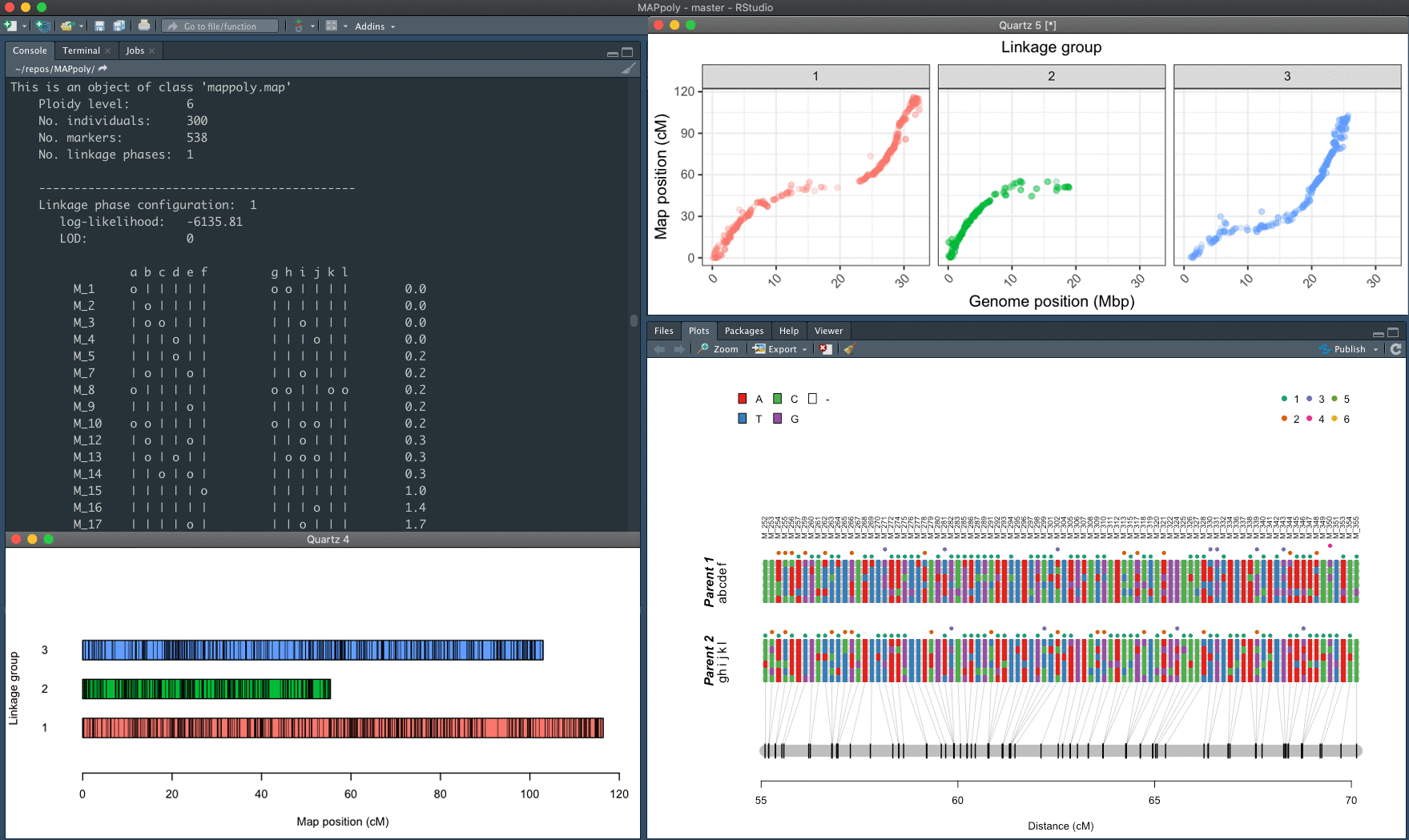
The mapping strategy is based on using pairwise recombination fraction estimation as the first source of information to position allelic variants in specific homologues sequentially. For situations where pairwise analysis has limited power, the algorithm relies on the multilocus likelihood obtained through a hidden Markov model (HMM). The derivation of the HMM used in MAPpoly can be found in Mollinari and Garcia, 2019.
To install MAPpoly from the The Comprehensive R Archive Network (CRAN) use
install.packages("mappoly")You can install the development version from Git Hub. Within R, you
need to install devtools:
install.packages("devtools")If you are using Windows, you must install the the latest recommended version of Rtools.
To install MAPpoly from Git Hub use
devtools::install_github("mmollina/mappoly", dependencies=TRUE)For further QTL analysis, we recommend our QTLpoly package. QTLpoly is an under development software to map quantitative trait loci (QTL) in full-sib families of outcrossing autopolyploid species based on a random-effect multiple QTL model Pereira et al. 2020.
R vignette("mappoly_startguide")# Enable this universe
options(repos = c(
polyploids = 'https://polyploids.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
# Install some packages
install.packages('mappoly')This package has been developed as part of the Genomic Tools for Sweetpotato Improvement project (GT4SP) and SweetGAINS, both funded by Bill & Melinda Gates Foundation. Its continuous improvement is made possible by Tools for polyploids, funded by USDA NIFA Specialty Crop Research Initiative Award.
<a id="NCSU" href="https://www.ncsu.edu/"><img src="https://brand.ncsu.edu/assets/logos/ncstate-brick-2x2-red.png" width="150" alt=""/></a>
<a id="BMGF" href="https://www.gatesfoundation.org/"><img src="https://fsm-alliance.org/wp-content/uploads/gates-logo-bda5cc0866e8e37eccab4ac502b916c1-copy.png" width="150" alt=""/></a>
<a id="GT4SP" href="https://sweetpotatogenomics.cals.ncsu.edu/"><img src="http://www.sweetpotatoknowledge.org/wp-content/uploads/2016/02/GT4SP-logo-e1456736272456.png" width="70" alt=""/></a>
<a id="sweetgains" href="https://cgspace.cgiar.org/handle/10568/106838"><img src="https://cipotato.org/wp-content/uploads/2020/06/SweetGains-sin-fondo-1-350x230.png" width="150" alt=""/></a>
<a id="PolyploidTools" href="https://www.polyploids.org/"><img src="https://www.polyploids.org/sites/default/files/inline-images/Project%20Logo-transparent.png" width="180" alt=""/></a>
<a id="USDA-NIFA" href="https://nifa.usda.gov/"><img src="https://upload.wikimedia.org/wikipedia/commons/0/06/USDA_NIFA_Twitter_Logo.jpg" width="100" alt=""/></a>
<span class="stretch"></span>NC State University promotes equal opportunity and prohibits discrimination and harassment based upon one’s age, color, disability, gender identity, genetic information, national origin, race, religion, sex (including pregnancy), sexual orientation and veteran status.