

You can access the CheatSheet for overviewR here
The goal of overviewR is to make it easy to get an overview of a data set by displaying relevant sample information. At the moment, there are the following functions:
overview_tab generates a tabular overview of the
sample. The general sample plots a two-column table that provides
information on an id in the left column and a the time frame on the
right column.overview_crosstab generates a cross table. The
conditional column allows to disaggregate the overview table by
specifying two conditions, hence resulting a 2x2 table. This way, it is
easy to visualize the time and scope conditions as well as theoretical
assumptions with examples from the data set.overview_print converts the output of both
overview_tab and overview_crosstab into LaTeX
code and/or directly into a .tex file.overview_plot is an alternative to visualize the sample
(a way to present results from overview_tab)overview_crossplot is an alternative to visualize a
cross table (a way to present results from
overview_crosstab)overview_heat plots a heat map of your time lineoverview_na plots an overview of missing values by
variableoverview_overlap plots comparison plots (bar graph and
Venn diagram) to compare to data framesThe plots can be saved using the ggsave() command. The
output of overview_tab and overview_crosstab
are also compatible with other packages such as xtable,
flextable,
or knitr.
We present a short step-by-step guide as well as the functions in more detail below.
A stable version of overviewR can be directly accessed
on CRAN:
install.packages("overviewR", force = TRUE)To install the latest development version of overviewR
directly from GitHub use:
library(devtools) # Tools to Make Developing R Packages Easier # Tools to Make Developing R Packages Easier
devtools::install_github("cosimameyer/overviewR")First, load the package.
library(overviewR) # Easily Extracting Information About Your Data # Easily Extracting Information About Your DataThe following examples use a toy data set (toydata) that
comes with the package. This data contains artificially generated
information in a cross-sectional format on 5 countries, covering the
period 1990-1999.
data(toydata)
head(toydata)
#> ccode year month gdp population
#> 1 RWA 1990 Jan 24180.77 14969.988
#> 2 RWA 1990 Feb 23650.53 11791.464
#> 3 RWA 1990 Mar 21860.14 30047.979
#> 4 RWA 1990 Apr 20801.06 19853.556
#> 5 RWA 1990 May 18702.84 5148.118
#> 6 RWA 1990 Jun 30272.37 48625.140There are 264 observations for 5 countries (Angola, Benin, France,
Rwanda, and UK) stored in the ccode variable, over a time
period between 1990 to 1999 (year) with additional
information for the month (month). Additionally, two
artificially generated fake variables for GDP (gdp) and
population size (population) are included to illustrate of
conditions.
The following functions work best on data sets that have an
id-time-structure, in the case of toydata this corresponds
to country-year with ccode and year. If the
data set does not have this format yet, consider using pivot_wider()
or pivot_longer() to get to the format.
overview_tabGenerate some general overview of the data set using the time and
scope conditions with overview_tab.
output_table <- overview_tab(dat = toydata, id = ccode, time = year)The resulting data frame collapses the time condition for each id by
taking into account potential gaps in the time frame. Note that the
column name for the time frame is set by default to
time_frame and internally generated when using
overview_tab.
output_table# ccode time_frame
# RWA 1990 - 1995
# AGO 1990 - 1992
# BEN 1995 - 1999
# GBR 1991, 1993, 1995, 1997, 1999
# FRA 1993, 1996, 1999overview_crosstabTo generate a cross table that divides the data based on two
conditions, for instance GDP and population size,
overview_crosstab can be used. threshold1 and
threshold2 thereby indicate the cut point for the two
conditions (cond1 and cond2),
respectively.
output_crosstab <- overview_crosstab(
dat = toydata,
cond1 = gdp,
cond2 = population,
threshold1 = 25000,
threshold2 = 27000,
id = ccode,
time = year
)The data frame output looks as follows:
# part1 part2
# 1 AGO (1990, 1992), FRA (1993), GBR (1997) BEN (1996, 1999), FRA (1999), GBR (1993), RWA (1992, 1994)
# 2 BEN (1997), RWA (1990) AGO (1991), BEN (1995, 1998), FRA (1996), GBR (1991, 1995, 1999), RWA (1991, 1993, 1995)Note, if a data set is used that has multiple observations on the
id-time unit, the function automatically aggregates the data set using
the mean of condition 1 (cond1) and condition 2
(cond2).
overview_printTo generate an easily usable LaTeX output for the generated
overview_tab and overview_crosstab objects,
overviewR offers the function overview_print.
The following illustrate this using the output_table object
from overview_tab.
overview_print(obj = output_table)% Overview table generated in R version 4.0.0 (2020-04-24) using overviewR
% Table created on 2020-06-21
\begin{table}[ht]
\centering
\caption{Time and scope of the sample}
\begin{tabular}{ll}
\hline
Sample & Time frame \\
\hline
RWA & 1990 - 1995 \\
AGO & 1990 - 1992 \\
BEN & 1995 - 1999 \\
GBR & 1991, 1993, 1995, 1997, 1999 \\
FRA & 1993, 1996, 1999 \\
\hline
\end{tabular}
\end{table}
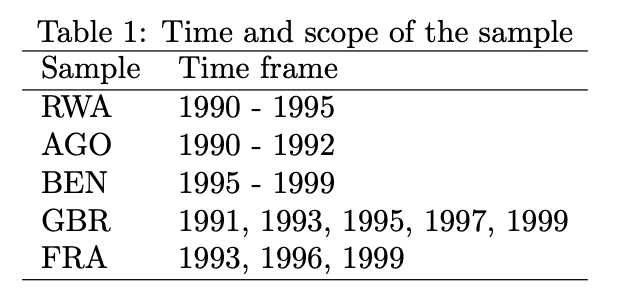
The default already provides a title (“Time and scope of the sample”)
that can be modified in the argument title. The same holds
for the column names (“Sample” and “Time frame” are set by default but
can be modified as shown below).
overview_print(obj = output_table, id = "Countries", time = "Years",
title = "Cool new title for our awesome table")% Overview table generated in R version 4.0.0 (2020-04-24) using overviewR
% Table created on 2020-06-21
\begin{table}[ht]
\centering
\caption{Cool new title for our awesome table}
\begin{tabular}{ll}
\hline
Countries & Years \\
\hline
RWA & 1990 - 1995 \\
AGO & 1990 - 1992 \\
BEN & 1995 - 1999 \\
GBR & 1991, 1993, 1995, 1997, 1999 \\
FRA & 1993, 1996, 1999 \\
\hline
\end{tabular}
\end{table}
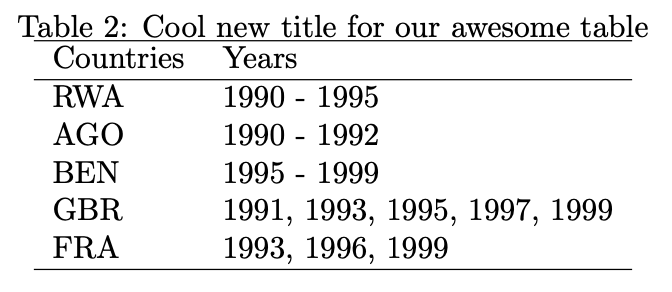
The same function can also be used for outputs from the
overview_crosstab function by using the argument
crosstab = TRUE. There are also options to label the
respective conditions (cond1 and cond2). Note
that this should correspond to the conditions (cond1 and
cond2) specified in the overview_crosstab
function.
overview_print(
obj = output_crosstab,
title = "Cross table of the sample",
crosstab = TRUE,
cond1 = "GDP",
cond2 = "Population"
)% Overview table generated in R version 4.0.0 (2020-04-24) using overviewR
% Table created on 2020-06-21
% Please add the following packages to your document preamble:
% \usepackage{multirow}
% \usepackage{tabularx}
% \newcolumntype{b}{X}
% \newcolumntype{s}{>{\hsize=.5\hsize}X}
\begin{table}[ht]
\caption{Cross table of the sample}
\begin{tabularx}{\textwidth}{ssbb}
\hline & & \multicolumn{2}{c}{\textbf{GDP}} \\
& & \textbf{Fulfilled} & \textbf{Not fulfilled} \\
\hline \\
\multirow{2}{*}{\textbf{Population}} & \textbf{Fulfilled} &
AGO (1990, 1992), FRA (1993), GBR (1997) & BEN (1996, 1999), FRA (1999), GBR (1993), RWA (1992, 1994)\\
\\ \hline \\
& \textbf{Not fulfilled} & BEN (1997), RWA (1990) & AGO (1991), BEN (1995, 1998), FRA (1996), GBR (1991, 1995, 1999), RWA (1991, 1993, 1995)\\ \hline \\
\end{tabularx}
\end{table}
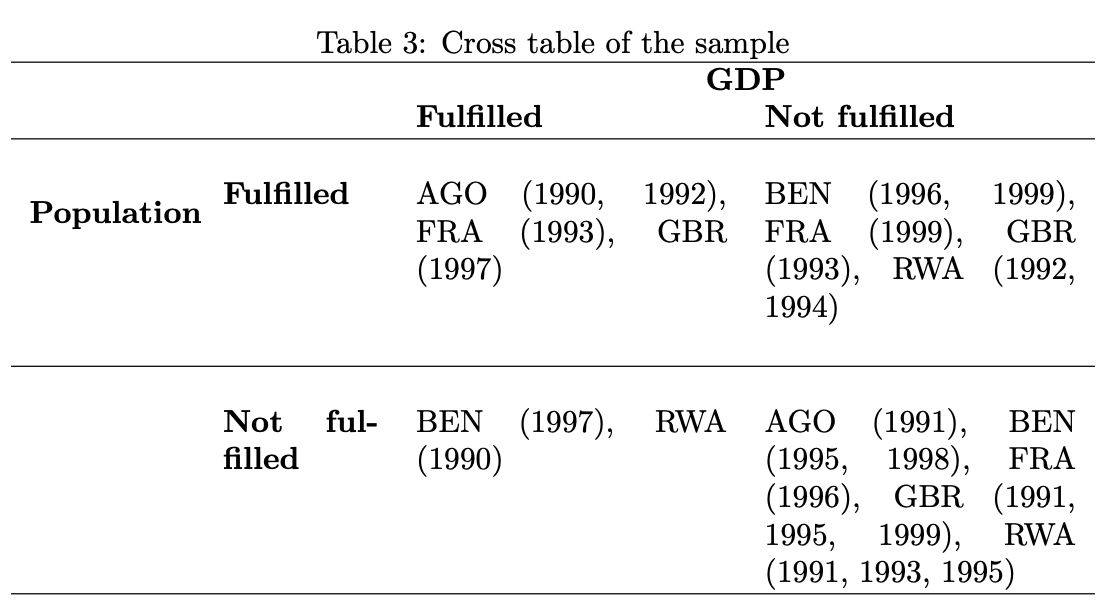
overview_print further allows more specifications such
as the font size or a a label. These functions are currently
supported only in the development version of the package.
overview_print(obj = output_table,
fontsize = "scriptsize",
label = "tab:overview")With save_out = TRUE the function exports the output as
a .tex file and stores it on the device.
overview_print(obj = output_table, save_out = TRUE, path = "SET-YOUR-PATH",
file = "output.tex")overview_plotIn addition to tables, overviewR also provides plots to
illustrate the structure of your data. overview_plot
illustrates the information that is generated in
overview_table in a ggplot graphic. All scope objects
(e.g., countries) are listed on the y-axis where horizontal lines
indicate the coverage across the entire time frame of the data (x-axis).
This helps to spot gaps in the data for specific scope objects and
outlines at what time point they occur.
data(toydata)
overview_plot(dat = toydata, id = ccode, time = year)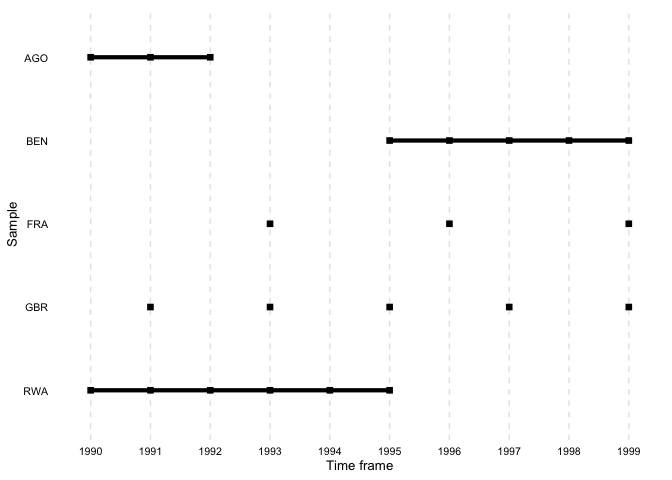
The results are sorted alphabetically by default. The order can also
be reversed by setting asc to FALSE.
overview_plot(dat = toydata, id = ccode, time = year, asc = FALSE)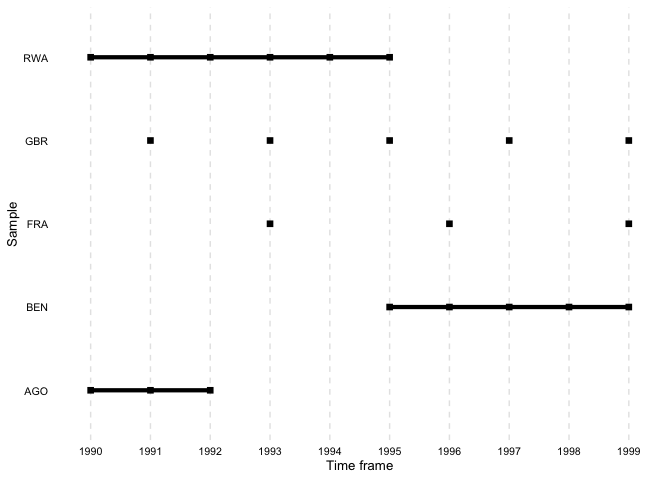
There is also an option to color the time lines conditionally. Here,
we introduce a dummy variable that indicates whether the year was before
1995 or not. We use this dummy to color the time lines using the
color argument. Note, this argument is currently only
implemented in the development version that can be accessed from
GitHub.
# Load the GitHub version
library(devtools) # Tools to Make Developing R Packages Easier
devtools::install_github("cosimameyer/overviewR")
#>
#> checking for file ‘/private/var/folders/23/l11m8s8s42x85pmh6100kf180000gn/T/Rtmpn9zPqU/remotesbc8c1543222f/cosimameyer-overviewR-c07706e/DESCRIPTION’ ... ✓ checking for file ‘/private/var/folders/23/l11m8s8s42x85pmh6100kf180000gn/T/Rtmpn9zPqU/remotesbc8c1543222f/cosimameyer-overviewR-c07706e/DESCRIPTION’
#> ─ preparing ‘overviewR’:
#> checking DESCRIPTION meta-information ... ✓ checking DESCRIPTION meta-information
#> ─ checking for LF line-endings in source and make files and shell scripts
#> ─ checking for empty or unneeded directories
#> ─ building ‘overviewR_0.0.7.999.1.tar.gz’
#>
#>
library(overviewR) # Easily Extracting Information About Your Data
library(magrittr) # A Forward-Pipe Operator for R
# Code whether a year was before 1995
toydata %<>%
dplyr::mutate(before = ifelse(year < 1995, 1, 0))
# Plot using the `color` argument
overview_plot(dat = toydata, id = ccode, time = year, color = before)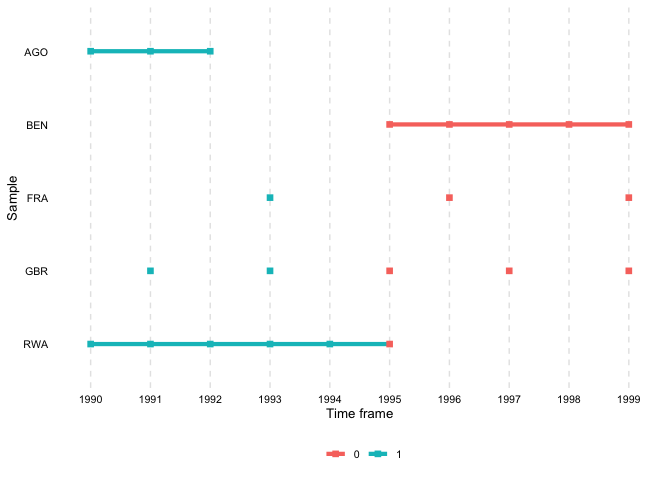
The development version also allows to change the dot size using the
dot_size argument. The default is “2”. Note, this
argument is currently only implemented in the development version that
can be accessed from GitHub.
# Plot using the `color` argument
overview_plot(dat = toydata, id = ccode, time = year, dot_size = 5)
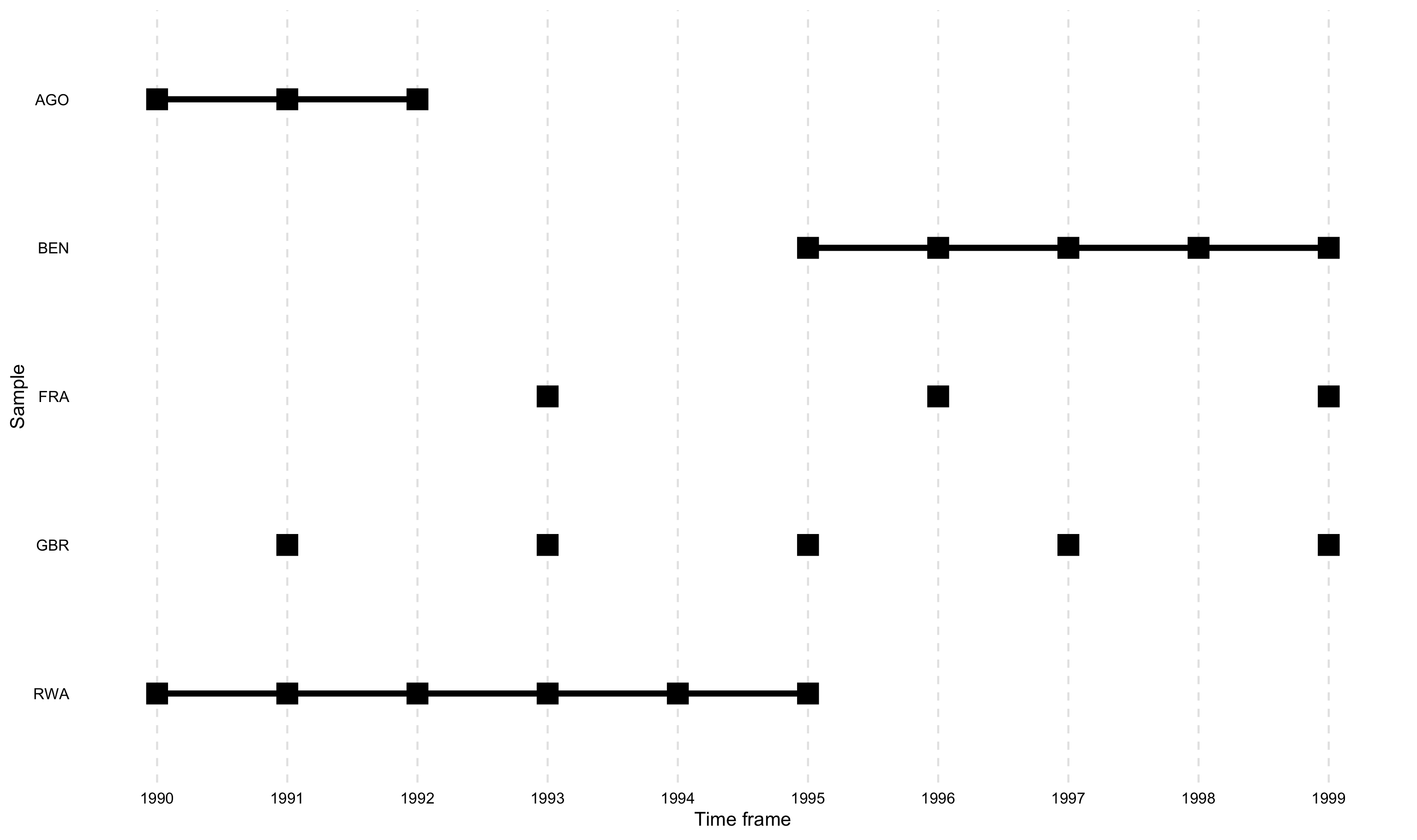
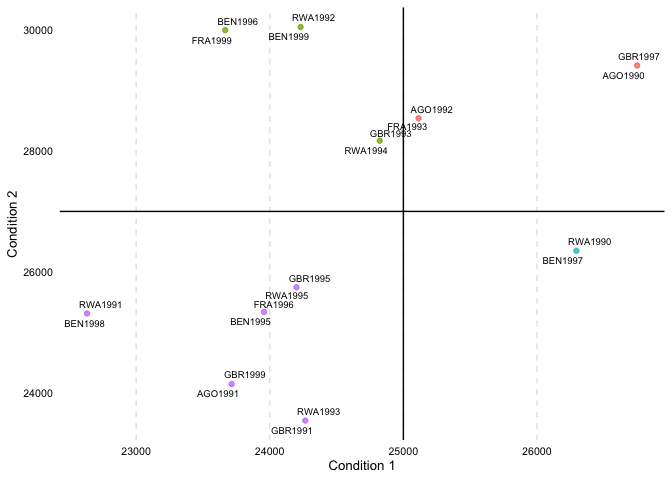
overview_crossplotTo visualize also the cross table, overview_crossplot
does the job. Note, this function is currently only implemented in
the development version that can be accessed from GitHub.
# Load the GitHub version
library(devtools) # Tools to Make Developing R Packages Easier
devtools::install_github("cosimameyer/overviewR")
library(overviewR) # Easily Extracting Information About Your Data
overview_crossplot(
toydata,
id = ccode,
time = year,
cond1 = gdp,
cond2 = population,
threshold1 = 25000,
threshold2 = 27000,
color = TRUE,
label = TRUE
)
overview_heatoverview_heat takes a closer look at the time and scope
conditions by visualizing the data coverage for each time and scope
combination in a ggplot heat map. This function is best explained using
an example. Suppose you have a dataset with monthly data for different
countries and want to know if data is available for each country in
every month. overview_heat intuitively does this by
plotting a heat map where each cell indicates the coverage for that
specific combination of time and scope (e,g., country-year). As
illustrated below, the darker the cell is, the more coverage it has. The
plot also indicates the relative or absolute coverage of each cell. For
instance, Angola (“AGO”) in 1991 shows the coverage of 75%. This means
that of all potential 12 months of coverage (12 months for one year),
only 9 are covered.
overview_heat(toydata_red,
ccode,
year,
perc = TRUE,
exp_total = 12)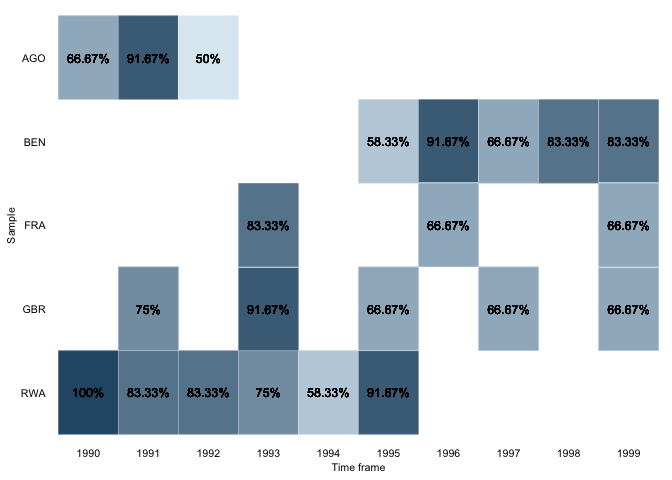
overview_naoverview_na is a simple function that provides
information about the content of all variables in your data, not only
the time and scope conditions. It returns a horizontal ggplot bar plot
that indicates the amount of missing data (NAs) for each variable (on
the y-axis). You can choose whether to display the relative amount of
NAs for each variable in percentage (the default) or the total number of
NAs.
overview_na(toydata_with_na)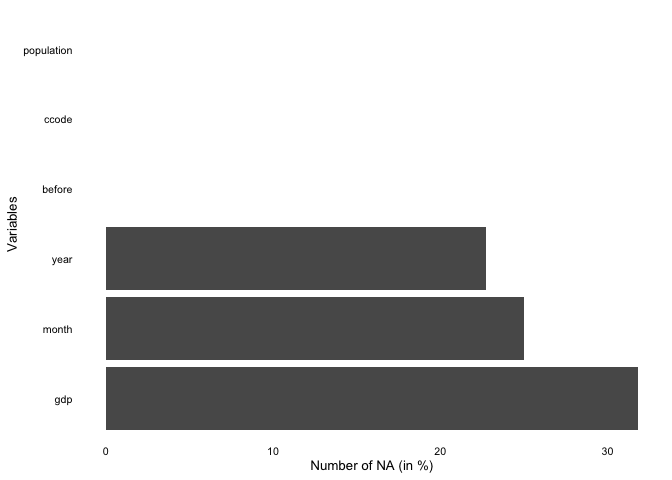
overview_na(toydata_with_na, perc = FALSE)
overview_overlapThis function allows to compare two data sets. We are currently working on an extended version that allows comparing >2 data sets. Note, this function is currently only implemented in the development version that can be accessed from GitHub.
At the current development stage, the function works as follows:
library(dplyr)
# Subset one data set for comparison
toydata2 <- toydata %>% dplyr::filter(year > 1992)
overview_overlap(
dat1 = toydata,
dat2 = toydata2,
dat1_id = ccode,
dat2_id = ccode,
plot_type = "bar" # This is the default
)
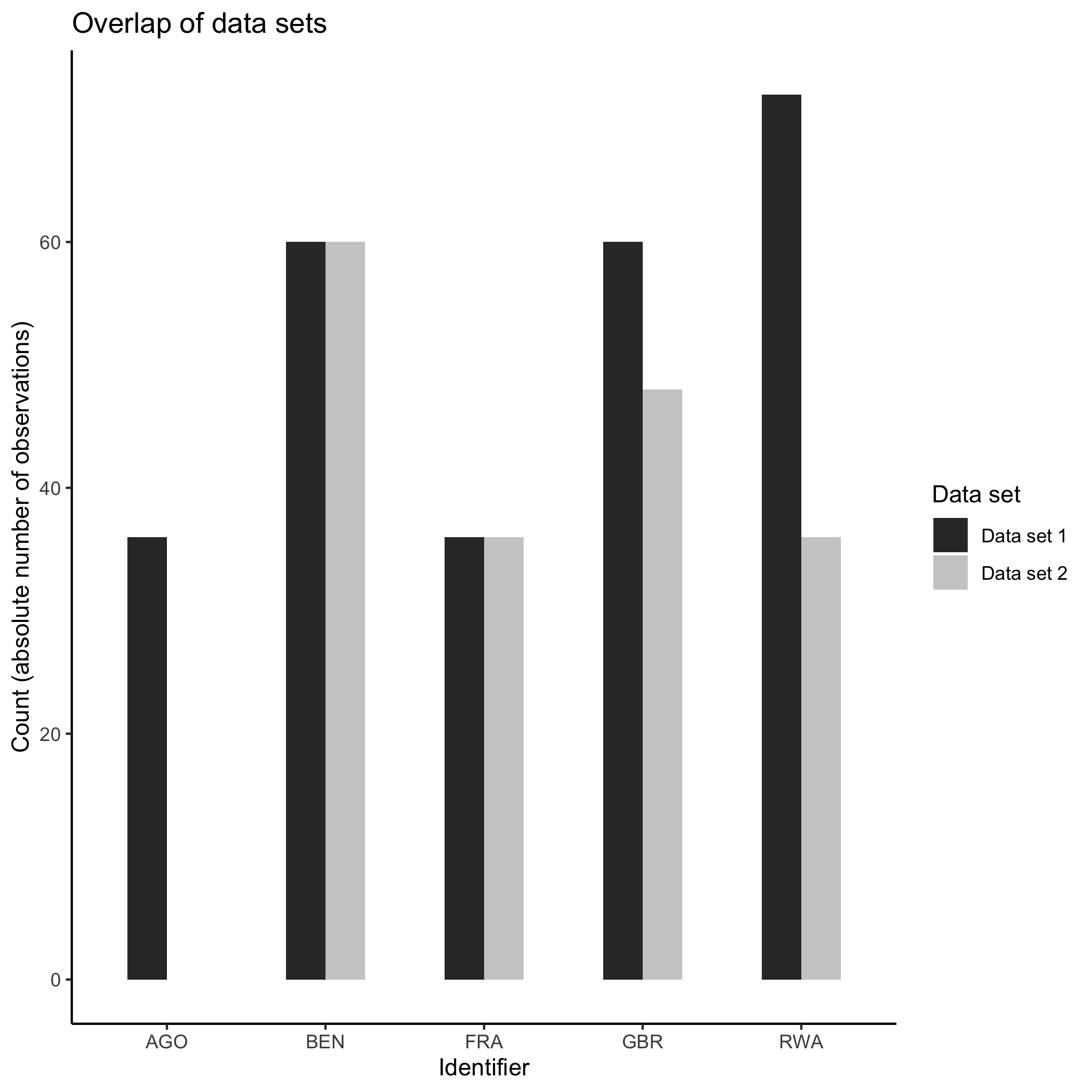
Or a Venn diagram
overview_overlap(
dat1 = toydata,
dat2 = toydata2,
dat1_id = ccode,
dat2_id = ccode,
plot_type = "venn"
)
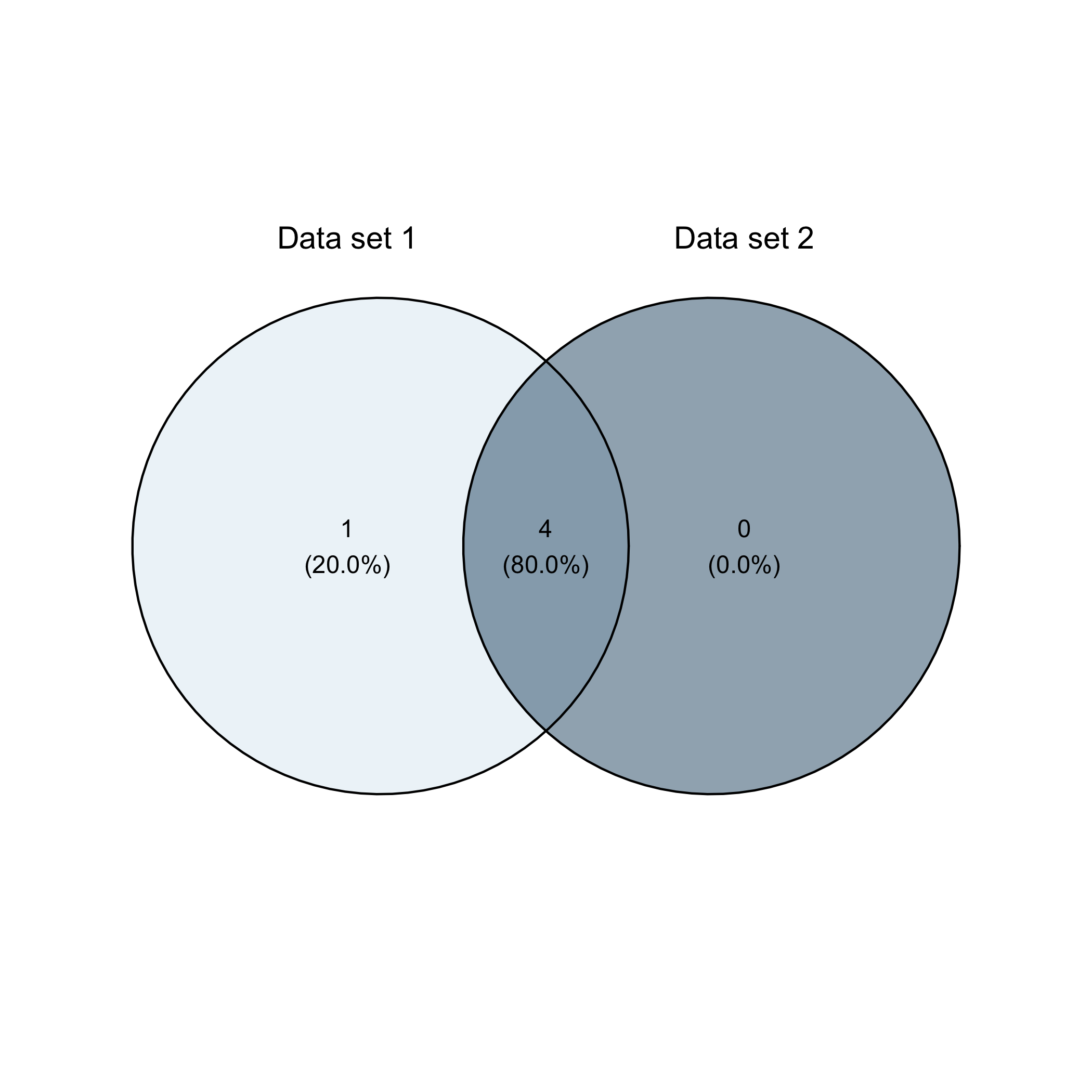
flextable, xtable, and kableThe outputs of overview_tab and
overview_crosstab are also compatible with other functions
such as xtable,
flextable,
or kable
from knitr.
Two examples are shown below:
library(flextable) # not installed on this machine
table_output <- qflextable(output_table)
table_output <-
set_header_labels(table_output,
ccode = "Countries",
time_frame = "Time frame")
set_table_properties(
table_output,
width = .4,
layout = "autofit"
)library(knitr) # A General-Purpose Package for Dynamic Report Generation in R
knitr::kable(output_table)| ccode | time_frame |
|---|---|
| RWA | 1990-1995 |
| AGO | 1990-1992 |
| BEN | 1995-1999 |
| GBR | 1991, 1993, 1995, 1997, 1999 |
| FRA | 1993, 1996, 1999 |
ggplot2 and other packagesThe plot functions are fully ggplot2 based. While a
theme is pre-defined, this can easily be overwritten.
A classical ggplot2 theme alternative
library(ggplot2) # Create Elegant Data Visualisations Using the Grammar of Graphics
overview_na(toydata_with_na) +
ggplot2::theme_minimal() 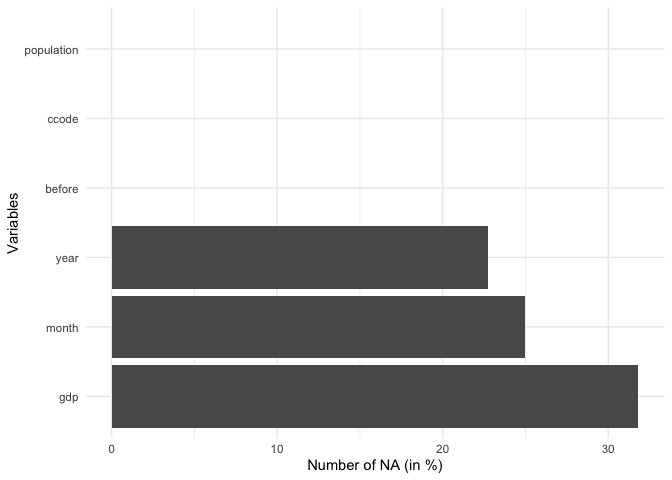
tidyverseAll functions are further easily accessible using a common
tidyverse workflow. Here are just three examples – the
possibilities are endless.
Using a filter function
library(dplyr) # A Grammar of Data Manipulation # A Grammar of Data Manipulation
toydata_with_na %>%
dplyr::filter(year > 1993) %>%
overview_na()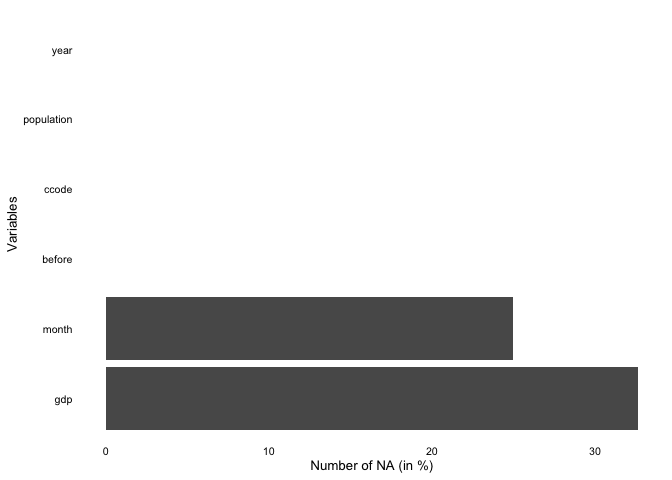
Using mutate to generate meaningful country names
library(countrycode) # Convert Country Names and Country Codes
library(dplyr) # A Grammar of Data Manipulation # A Grammar of Data Manipulation
toydata %>%
# Transform the country code (ISO3 character code) into a country name using the `countrycode` package
dplyr::mutate(country = countrycode::countrycode(ccode, "iso3c", "country.name")) %>%
overview_plot(id = country, time = year)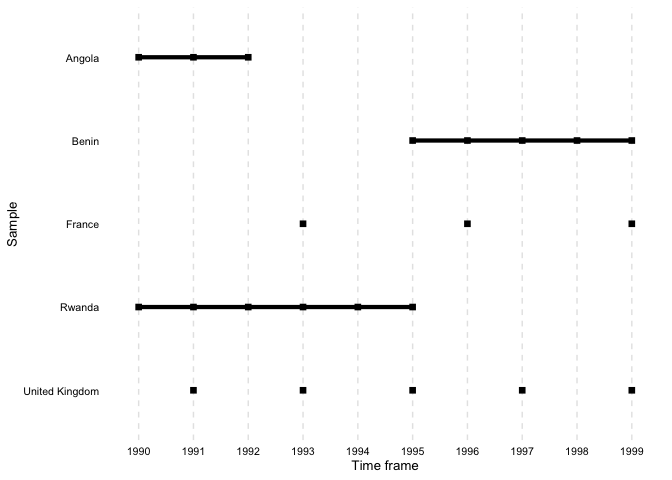
Using different overviewR functions after each other to
generate a workflow
# Produces a printable LaTeX output
toydata %>%
overview_tab(id = ccode, time = year) %>%
overview_print()% Overview table generated in R version 4.0.2 (2020-06-22) using overviewR
% Table created on 2020-12-30
\begin{table}[ht]
\centering
\caption{Time and scope of the sample}
\label{tab:tab1}
\begin{tabular}{ll}
\hline
Sample & Time frame \\
\hline
AGO & 1990 - 1992 \\
BEN & 1995 - 1999 \\
FRA & 1993, 1996, 1999 \\
GBR & 1991, 1993, 1995, 1997, 1999 \\
RWA & 1990 - 1995 \\
\hline
\end{tabular}
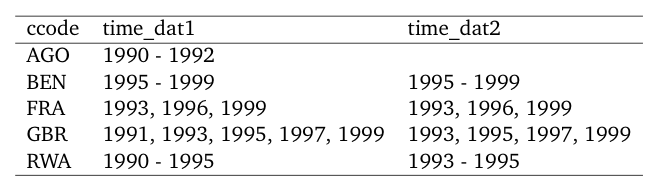
\end{table} If you wish to compare two data sets using overview_tab,
this is not (yet) implemented in overviewR but there is
currently a workaround.
library(overviewR)
library(dplyr)
library(xtable)
# Load data
data(toydata)
# Restrict the data so that we have something to compare :-)
toydata_res <- toydata %>%
dplyr::filter(year > 1992)
# Generate two overview_tab objects
dat1 <- overview_tab(toydata, id = ccode, time = year)
dat2 <- overview_tab(toydata_res, id = ccode, time = year)
# And now we use full_join to combine both
dat_full <- dat1 %>%
dplyr::full_join(dat2, by = "ccode") %>%
dplyr::rename(time_dat1 = time_frame.x,
time_dat2 = time_frame.y)Having a look at the output, we see that this is exactly what we wanted to have:
head(dat_full)#> # A tibble: 5 x 3
#> # Groups: ccode [5]
#> ccode time_dat1 time_dat2
#> <chr> <chr> <chr>
#> 1 AGO 1990 - 1992 <NA>
#> 2 BEN 1995 - 1999 1995 - 1999
#> 3 FRA 1993, 1996, 1999 1993, 1996, 1999
#> 4 GBR 1991, 1993, 1995, 1997, 1999 1993, 1995, 1997, 1999
#> 5 RWA 1990 - 1995 1993 - 1995overview_print cannot handle this object (yet), so we
use xtable instead which gives us the LaTeX output.
print(xtable(dat_full), include.rownames=FALSE)% latex table generated in R 4.0.2 by xtable 1.8-4 package
% Tue Feb 16 18:20:51 2021
\begin{table}[ht]
\centering
\begin{tabular}{lll}
\hline
ccode & time\_dat1 & time\_dat2 \\
\hline
AGO & 1990 - 1992 & \\
BEN & 1995 - 1999 & 1995 - 1999 \\
FRA & 1993, 1996, 1999 & 1993, 1996, 1999 \\
GBR & 1991, 1993, 1995, 1997, 1999 & 1993, 1995, 1997, 1999 \\
RWA & 1990 - 1995 & 1993 - 1995 \\
\hline
\end{tabular}
\end{table}
The hex sticker is generated by ourselves using the hexSticker
package.