rtsVisA lightweight R package to visualize large raster time series, building on a fast temporal interpolation core. rtsVis is linked to the moveVis package and their joint use is recommended.

The released version of rtsVis can be installed from CRAN:
The latest version of the package can be installed from github.
rtsVis operates on lists of objects:
To process those lists in a pipeline, we recommend pipes such as provided by magrittr.
ts_raster Assemble/interpolate a raster time series.ts_fill_na Fill NA values in a raster time series ### Creating Framests_flow_frames Create a series of charts of a raster time series.ts_makeframes Create spatial ggplots of a raster time series.ts_add_positions_to_frames Add points, coordinates, or polygons to a list of spatial plots.In this example, we use a MODIS time series (download here)to create typical visualisations which highlight vegetation dynamics on the African continent.
We read our inputs:
Optional, but useful to enhance the visualizations:
Some functions in rtsVis require that timestamps for the rasters are set. Often, these come with the metadata or can be derived from the filename. They can also be manually set. In this example, we have monthly medians, and no true acquisition time. Therefore, we manually create a series of dates. In addition to the input times, we also set output times - for these, output dates will be created. We simply create a second, denser series of dates. Having a denser series will smooth the animation.
If images from the input series are missing, it is not an issue, at least not from the technical side of things. Images for out_times will be interpolated regardless of the regularity of the input data. To illustrate this, we remove one of the input images.
library(raster)
library(sf)
library(rtsVis)
library(RStoolbox)
library(tidyverse)
# Load the inputs
modis_tifs <- list.files("Beispieldaten/MODIS/Africa_MODIS_2017-2020/",full.names = T,pattern=".tif$")
points <- st_read("Beispieldaten/Ancillary/Africa/Africa_places.gpkg")
outline <- st_read("Beispieldaten/Ancillary/Outline_continents_africa.gpkg")
hillshade <- raster("Beispieldaten/Ancillary/SR_LR/SR_LR.tif") %>% readAll()
#Create in-times and out-times
in_dates <- as.POSIXct(seq.Date(as.Date("2017-01-15"),
as.Date("2020-12-15"),
length.out = length(modis_filled)))
out_dates <-seq.POSIXt(from = in_dates[1],
to = in_dates[length(in_dates)],
length.out = length(in_dates)*4 )
# simulate a missing image
in_dates <- in_dates[-13]
modis_tifs <- modis_tifs[-13]For the preparation of the rasters, we use three functions:
stack A raster function which we use with lapply to load the tifs into a list.ts_fill_na We fill, wherever possible, missing values using simple temporal interpolation. This is not strictly necessary, but improves the visualization.ts_raster We assemble a full time series. This will interpolate additional missing frames for every desired output date and can take a long time for large time-series.Often, data requires additional preprocessing steps, such as reprojection, cropping, or masking. These can be applied with lapply to retain the list format.
ts_raster returns a list of interpolated rasters, one for each desired output date, with timestamps attached.
#Read the images as stacks
modis_tifs_stacks <- modis_tifs %>% lapply(stack)
#fill NAs
modis_filled <- ts_fill_na(modis_tifs_stacks)
modis_ts <- ts_raster(
r_list = modis_filled,
r_times = in_dates,
out_times = out_dates,
fade_raster = T) In this step, ts_makeframes is used to create a list of frames (ggplot objects) from the time series (raster objects). We also add a outline to the plot area. This is one example of adding a position to a time series. Using pipes is not necessary, but improves readability greatly.
#create the frames from the raster
modis_frames_RGB <-
ts_makeframes(x_list = modis_ts,
l_indices = c(1,4,3), # MODIS bands are Red, NIR, Blue, Green
minq = 0.0) # adjust the stretch slightly
# Add the outline of the continent for visual clarity
modis_frames_RGB_ol <-
modis_frames_RGB %>%
ts_add_positions_to_frames(
positions = outline,
psize = 1)
# Use moveVis utility to animate the frames into a gif
moveVis::animate_frames(modis_frames_RGB_ol,
overwrite = TRUE,
out_file = "modis_frames_RGB_ol.gif",
height=300,
width=300,
fps = 10,
res=75)

The animation created suggests notable vegetation dynamics. An easy way to highlight this is the NDVI.
We calculate the NDVI using the overlay function provided by the raster package, and reattach the timestamps using ts_copy_frametimes. Thereby we receive a second time series with a single layer per timestep. Thus, we do not create RGB frames, but gradient frames.
Note that the individual frames are simply ggplot objects. Hence, moveVis functions and ggplot functions can be easily applied to the created frames using lapply.
#calculate the NDVI per image
modis_ndvi <- modis_ts %>% lapply(function(x){
ndvi <- overlay(x[[2]], x[[1]], fun=function(nir,red){(nir-red)/(nir+red)})
}) %>% rtsVis::ts_copy_frametimes(modis_ts)
# Create the frames from the NDVI raster
modis_ndvi_fr <-
ts_makeframes(x_list = modis_ndvi,
hillshade = hillshade,
r_type = "gradient")
# The frames can be adjusted like any ggplot
modis_ndvi_fr_styled <- modis_ndvi_fr %>% lapply(FUN=function(x){
x+scale_fill_gradient2("NDVI",
low = "red",
mid="yellow",
high = "green4",
midpoint=0.3,
limits=c(-0,1),
na.value = NA)+
theme_bw() +
xlab("Longitude")+
ylab("Latitude")+
ggtitle("Seasonality of NDVI in Africa",
"Derived from MCD43A4 -- Monthly Composite 2015-2020 -- Projection: WGS 84 (EPSG 4326)")
}) %>%
# packages like moveVis provide additional mapping functions
moveVis::add_northarrow(colour = "black", position = "bottomleft") %>%
moveVis::add_progress()rtsVis uses vector objects (positions) in two different ways:
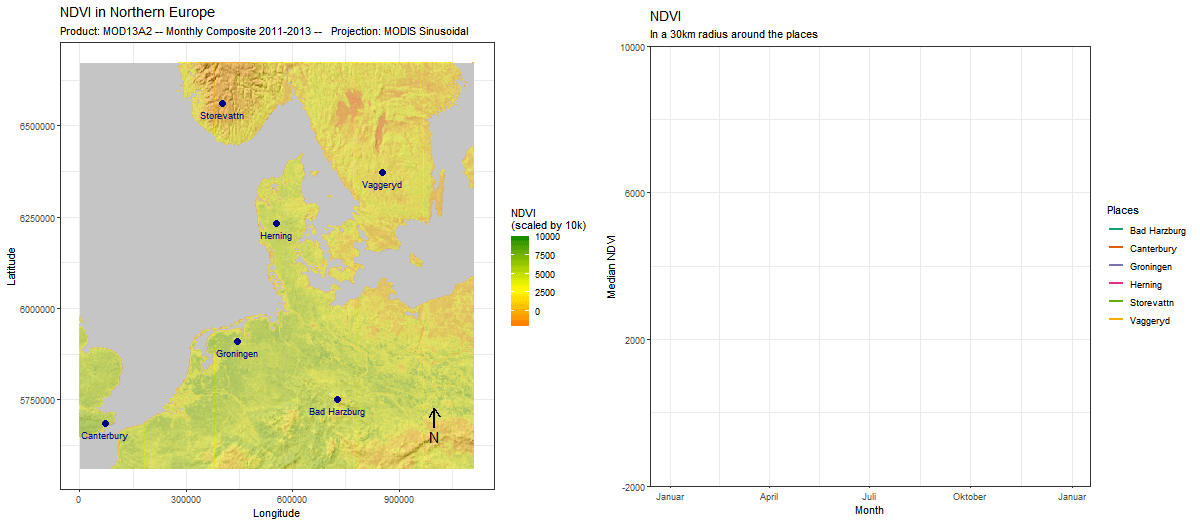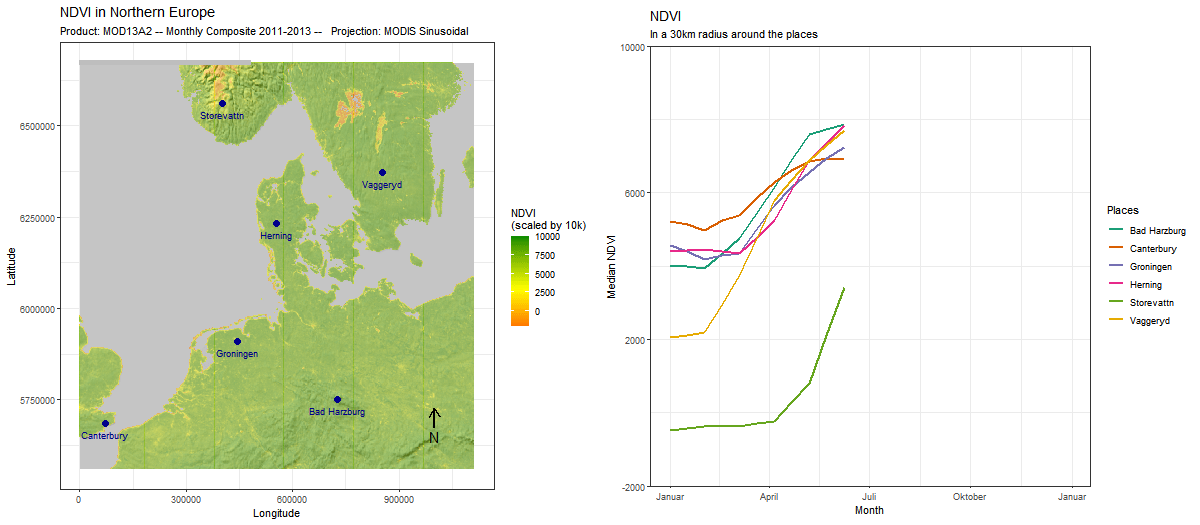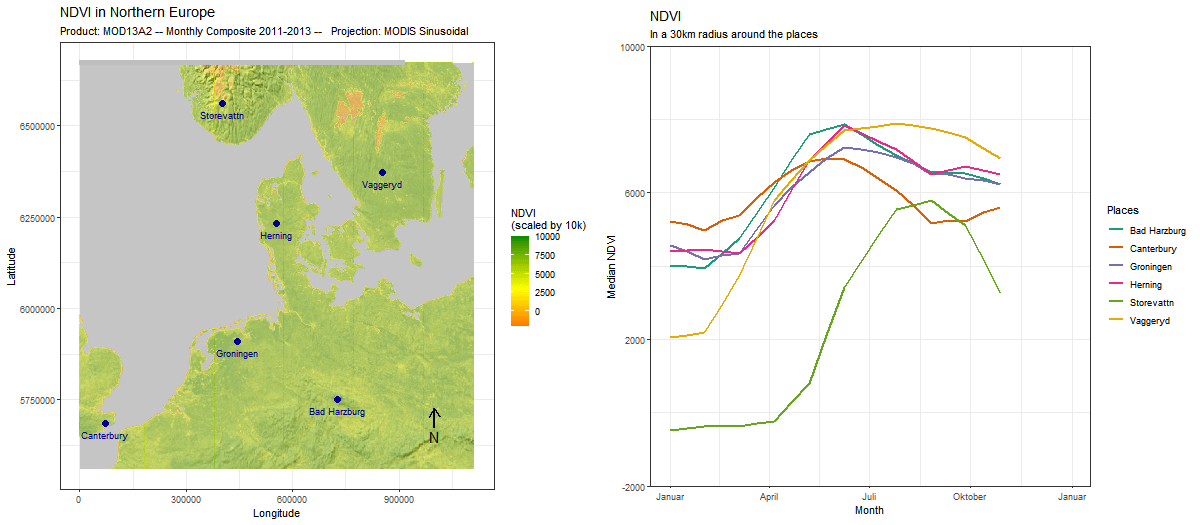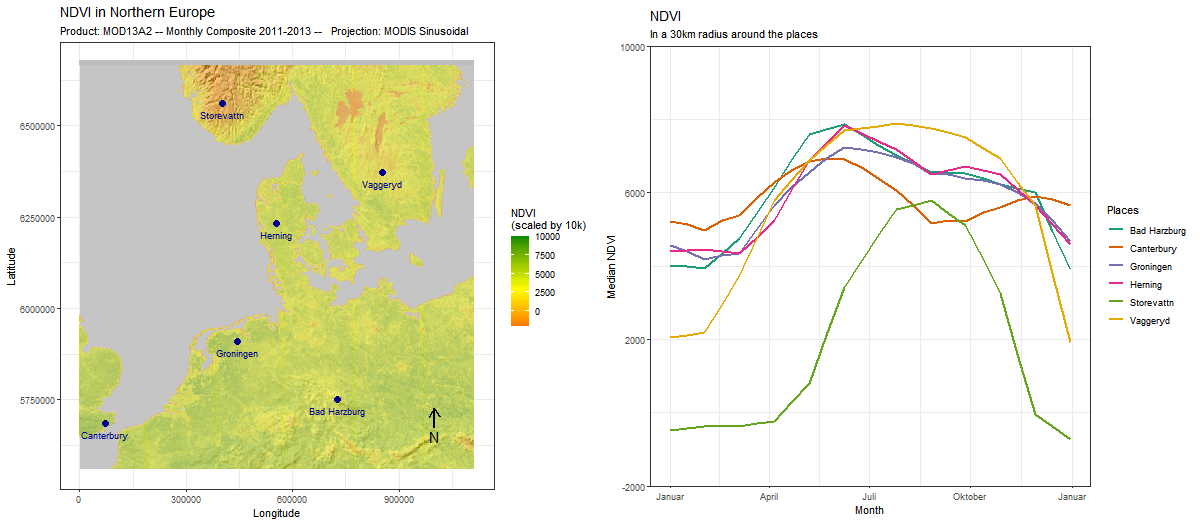
Sometimes, adding a position is for visual appeal only, as we do here by adding an outline to the continents. But often, it makes sense to use the two together. In this example, we use (ts_add_positions_to_frames) to add several locations as points to our spatial frames. Subsequently, we extract values in a buffer around these locations and create a line chart that visualizes the development of these values over time.
#reduce the number of points slightly
modis_ndvi_fr_styled_pos <-
modis_ndvi_fr_styled %>%
ts_add_positions_to_frames(outline,pcol = "white",psize = 1) %>%
ts_add_positions_to_frames(positions = points,
position_names = points$Name,
tcol = "blue4",
t_hjust = -1.5,
tsize = 4,
psize=3,
pcol="blue4",
add_text = T,col_by_pos = T)
# rtsVis comes with a variety of plotting functions. We select line2
# which maps color to position
modis_ndvi_lineframes <-
modis_ndvi %>%
ts_flow_frames(positions = points,
position_names = points$Name,
pbuffer = 0.1,
val_min = -0,
val_max = 1,
legend_position = "right",
plot_function = "line2",
aes_by_pos = TRUE,
position_legend = TRUE) %>%
# Flow frames too can be adjusted like any ggplot,
# for instance, to set a specific color scale
lapply(FUN = function(x){
x+
ggtitle("NDVI", "In a 1° radius around the places")+
ylab("Median NDVI")+
xlab("Year")+
theme(aspect.ratio = 0.3)+
scale_color_brewer("Places",palette = "Set1")
})Since the positions and the flow frames are thematically connected, it makes sense to combine the two in a single animation. For this, we again use moveVis functionalities.
Before we do that, we make sure that the colors of the points match those in the graph. Again, existing frames can be easily modified using lapply and ggplot2 syntax.
# apply the same color palette to the spatial frames
modis_ndvi_fr_styled_pos <- modis_ndvi_fr_styled_pos %>% lapply(function(x){
x+
scale_color_brewer("Places",palette = "Set1")
})
#Join and animate the frames using moveVis functionalities
modis_ndvi_joined <- moveVis::join_frames(
list(modis_ndvi_fr_styled,
modis_ndvi_lineframes)
)
moveVis::animate_frames(modis_ndvi_joined,
overwrite = TRUE,
out_file = "modis_frames_NDVI.gif",
height=525,
width=1200,
fps = 10,
res=75)
rtsVisprovides a number of preset plot types for visualising multispectral, gradient, or discrete rasters:
Line plots(line and line2)
Violin charts (vio)
Density charts (dens and dens2)
Pie charts (pie)
Bar charts (bar_stack and bar_fill)
They are all used in the same way, by passing the respective argument to ts_flow_frames:
# Violin frames visualising the distributions of the different bands at different positions
modis_ts_vioframes <-
modis_ts %>%
ts_flow_frames(positions = points[1:3,],
position_names = points[1:3,]$Name,
pbuffer = 3,
legend_position = "right",
plot_function = "vio",aes_by_pos = F,
position_legend = TRUE,band_colors = c("Red","Purple","Blue","Darkgreen"))
# Density frames visualizing the distributions of the NDVI across positions
modis_ts_densframes <-
modis_ndvi %>%
ts_flow_frames(positions = points[5:7,],
position_names = points[5:7,]$Name,
pbuffer = 3,
plot_function = "dens2",
band_legend_title = "NDVI",
band_names = "NDVI",band_colors = c("olivegreen"))
# plenty other types and custom plot functions...It is also possible to create custom plot functions and pass them to ts_flow_frames, like this:
custom_plot_function <- function(edf,pl,lp, bl, blt,plt, ps, vs,abp){
# ... Code for the creation of the ggplot
# return (plot)
}
modis_ts_vioframes <-
modis_ts %>%
ts_flow_frames(positions = points,
position_names = points$Name,
pbuffer = 3,
plot_function = custom_plot_function)

For more examples, and a guide on how to create custom plot functions, check out the Demo repository.
In development, published on CRAN. Last updated: 2021-05-18 17:30:00 CEST
To learn more about creating custom plot types, access the guide and test material at the associated github repo.
For creating visualization with movement data, visit moveVis.
For inspiration, visit the r-graph-gallery!
To learn about data visualization see Claus Wilke’s excellent book.